Structural Determinants of RGS-RhoGEF Signaling Critical to Entamoeba histolytica Pathogenesis.
Bosch, D.E., Kimple, A.J., Manning, A.J., Muller, R.E., Willard, F.S., Machius, M., Rogers, S.L., Siderovski, D.P.(2013) Structure 21: 65-75
- PubMed: 23260656
- DOI: https://doi.org/10.1016/j.str.2012.11.012
- Primary Citation of Related Structures:
4GOU - PubMed Abstract:
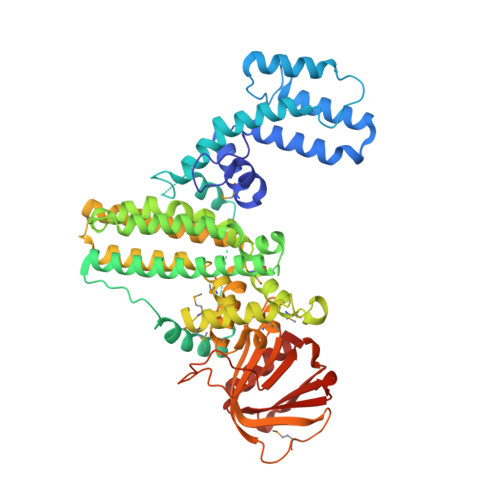
G protein signaling pathways, as key components of physiologic responsiveness and timing, are frequent targets for pharmacologic intervention. Here, we identify an effector for heterotrimeric G protein α subunit (EhGα1) signaling from Entamoeba histolytica, the causative agent of amoebic colitis. EhGα1 interacts with this effector and guanosine triphosphatase-accelerating protein, EhRGS-RhoGEF, in a nucleotide state-selective fashion. Coexpression of EhRGS-RhoGEF with constitutively active EhGα1 and EhRacC leads to Rac-dependent spreading in Drosophila S2 cells. EhRGS-RhoGEF overexpression in E. histolytica trophozoites leads to reduced migration toward serum and lower cysteine protease activity, as well as reduced attachment to, and killing of, host cells. A 2.3 Å crystal structure of the full-length EhRGS-RhoGEF reveals a putative inhibitory helix engaging the Dbl homology domain Rho-binding surface and the pleckstrin homology domain. Mutational analysis of the EhGα1/EhRGS-RhoGEF interface confirms a canonical "regulator of G protein signaling" domain rather than a RhoGEF-RGS ("rgRGS") domain, suggesting a convergent evolution toward heterotrimeric and small G protein cross-talk.
Organizational Affiliation:
Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.