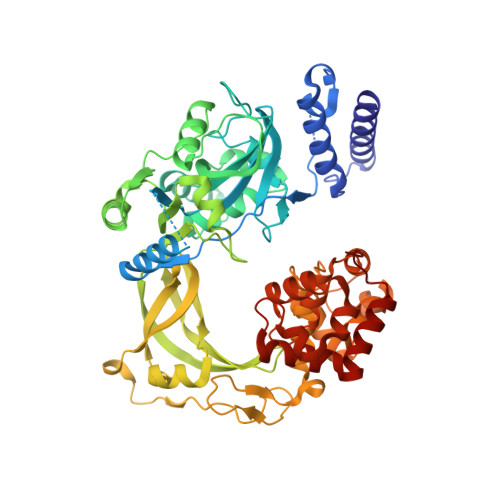
Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity.
Surivet, J.P., Lange, R., Hubschwerlen, C., Keck, W., Specklin, J.L., Ritz, D., Bur, D., Locher, H., Seiler, P., Strasser, D.S., Prade, L., Kohl, C., Schmitt, C., Chapoux, G., Ilhan, E., Ekambaram, N., Athanasiou, A., Knezevic, A., Sabato, D., Chambovey, A., Gaertner, M., Enderlin, M., Boehme, M., Sippel, V., Wyss, P.(2012) Bioorg Med Chem Lett 22: 6705-6711
- PubMed: 23006603
- DOI: https://doi.org/10.1016/j.bmcl.2012.08.094
- Primary Citation of Related Structures:
4GLW, 4GLX - PubMed Abstract:
A series of 2-amino-[1,8]-naphthyridine-3-carboxamides (ANCs) with potent inhibition of bacterial NAD(+)-dependent DNA ligases (LigAs) evolved from a 2,4-diaminopteridine derivative discovered by HTS. The design was guided by several highly resolved X-ray structures of our inhibitors in complex with either Streptococcus pneumoniae or Escherichia coli LigA. The structure-activity-relationship based on the ANC scaffold is discussed. The in-depth characterization of 2-amino-6-bromo-7-(trifluoromethyl)-[1,8]-naphthyridine-3-carboxamide, which displayed promising in vitro (MIC Staphylococcus aureus 1 mg/L) and in vivo anti-staphylococcal activity, is presented.
Organizational Affiliation:
Actelion Pharmaceuticals Ltd, Gewerbestrasse 16, CH-4123 Allschwil, Switzerland. jean-philippe.surivet@actelion.com