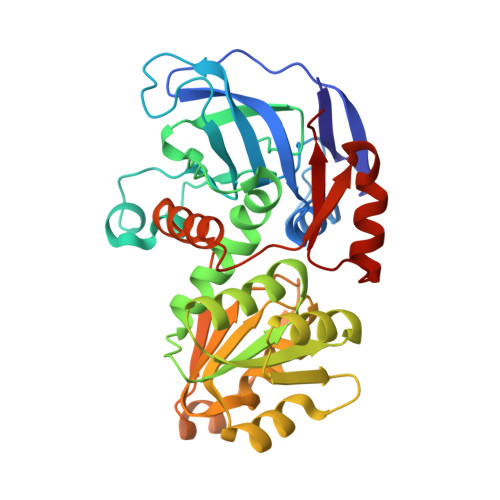
Structure of Escherichia coli AdhP (ethanol-inducible dehydrogenase) with bound NAD.
Thomas, L.M., Harper, A.R., Miner, W.A., Ajufo, H.O., Branscum, K.M., Kao, L., Sims, P.A.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 730-732
- PubMed: 23832197
- DOI: https://doi.org/10.1107/S1744309113015170
- Primary Citation of Related Structures:
4GKV - PubMed Abstract:
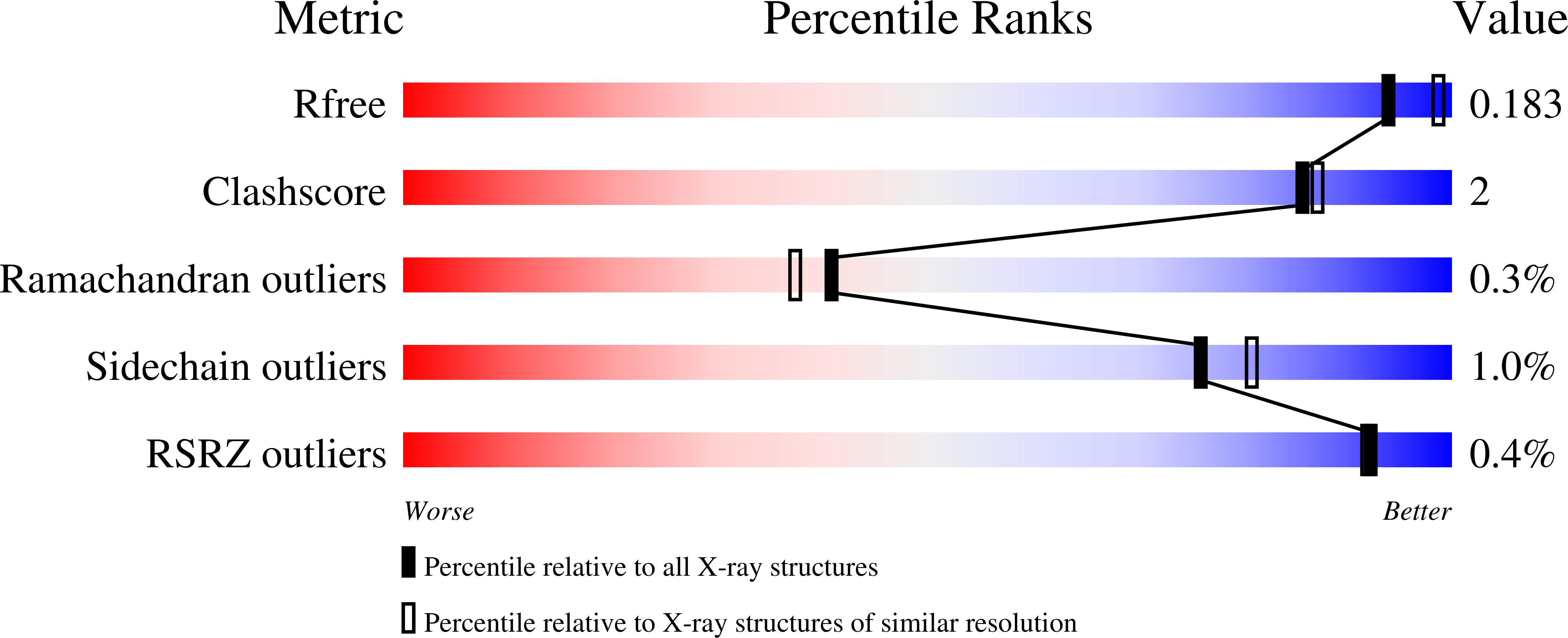
The crystal structure of AdhP, a recombinantly expressed alcohol dehydrogenase from Escherichia coli K-12 (substrain MG1655), was determined to 2.01 Å resolution. The structure, which was solved using molecular replacement, also included the structural and catalytic zinc ions and the cofactor nicotinamide adenine dinucleotide (NAD). The crystals belonged to space group P21, with unit-cell parameters a = 68.18, b = 118.92, c = 97.87 Å, β = 106.41°. The final R factor and Rfree were 0.138 and 0.184, respectively. The structure of the active site of AdhP suggested a number of residues that may participate in a proton relay, and the overall structure of AdhP, including the coordination to structural and active-site zinc ions, is similar to those of other tetrameric alcohol dehydrogenase enzymes.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Oklahoma, 101 Stephenson Parkway, Norman, OK 73019-5251, USA.