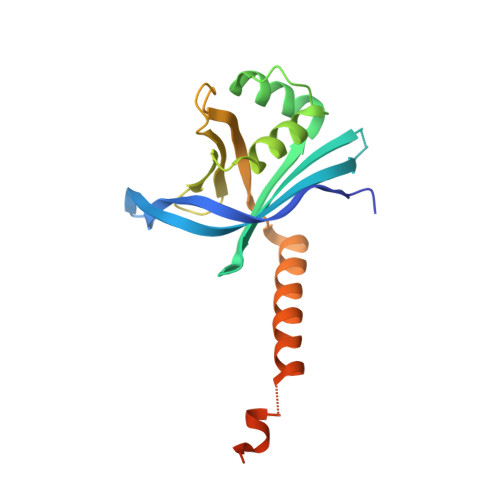
Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry.
Hilbert, M., Erat, M.C., Hachet, V., Guichard, P., Blank, I.D., Fluckiger, I., Slater, L., Lowe, E.D., Hatzopoulos, G.N., Steinmetz, M.O., Gonczy, P., Vakonakis, I.(2013) Proc Natl Acad Sci U S A 110: 11373-11378
- PubMed: 23798409
- DOI: https://doi.org/10.1073/pnas.1302721110
- Primary Citation of Related Structures:
4G79, 4GEU, 4GEX, 4GFA, 4GFC - PubMed Abstract:
Centrioles are evolutionary conserved organelles that give rise to cilia and flagella as well as centrosomes. Centrioles display a characteristic ninefold symmetry imposed by the spindle assembly abnormal protein 6 (SAS-6) family. SAS-6 from Chlamydomonas reinhardtii and Danio rerio was shown to form ninefold symmetric, ring-shaped oligomers in vitro that were similar to the cartwheels observed in vivo during early steps of centriole assembly in most species. Here, we report crystallographic and EM analyses showing that, instead, Caenorhabotis elegans SAS-6 self-assembles into a spiral arrangement. Remarkably, we find that this spiral arrangement is also consistent with ninefold symmetry, suggesting that two distinct SAS-6 oligomerization architectures can direct the same output symmetry. Sequence analysis suggests that SAS-6 spirals are restricted to specific nematodes. This oligomeric arrangement may provide a structural basis for the presence of a central tube instead of a cartwheel during centriole assembly in these species.
Organizational Affiliation:
Laboratory of Biomolecular Research (LBR), Paul Scherrer Institut, CH-5232 Villigen PSI, Switzerland.