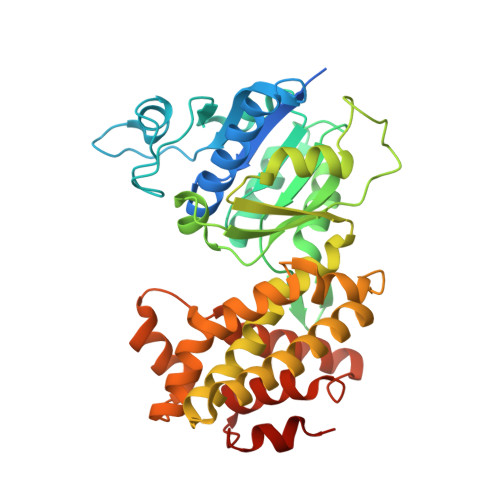
Structure of glycerol-3-phosphate dehydrogenase (GPD1) from Saccharomyces cerevisiae at 2.45A resolution
Alarcon, D.A., Nandi, M., Carpena, X., Fita, I., Loewen, P.C.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1279-1283
- PubMed: 23143232
- DOI: https://doi.org/10.1107/S1744309112037736
- Primary Citation of Related Structures:
4FGW - PubMed Abstract:
The interconversion of glycerol 3-phosphate and dihydroxyacetone phosphate by glycerol-3-phosphate dehydrogenases provides a link between carbohydrate and lipid metabolism and provides Saccharomyces cerevisiae with protection against osmotic and anoxic stress. The first structure of a glycerol-3-phosphate dehydrogenase from S. cerevisiae, GPD1, is reported at 2.45 Å resolution. The asymmetric unit contains two monomers, each of which is organized with N- and C-terminal domains. The N-terminal domain contains a classic Rossmann fold with the (β-α-β-α-β)2 motif typical of many NAD+-dependent enzymes, while the C-terminal domain is mainly α-helical. Structural and phylogenetic comparisons reveal four main structure types among the five families of glycerol-3-phosphate and glycerol-1-phosphate dehydrogenases and reveal that the Clostridium acetobutylican protein with PDB code 3ce9 is a glycerol-1-phosphate dehydrogenase.
Organizational Affiliation:
Institute for Research in Biomedicine, Parc Científic, Baldiri Reixac 10, 08028 Barcelona, Spain.