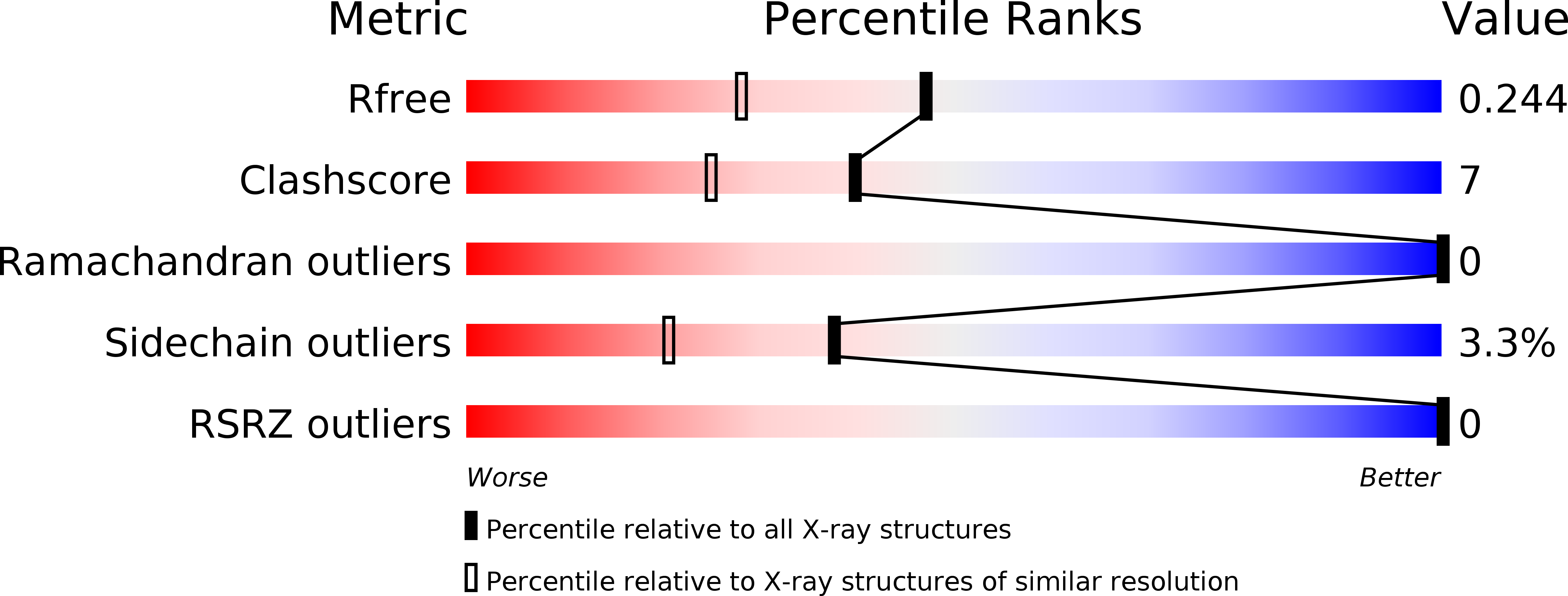
Structural basis of the novel S. pneumoniae virulence factor, GHIP, a glycosyl hydrolase 25 participating in host-cell invasion.
Niu, S., Luo, M., Tang, J., Zhou, H., Zhang, Y., Min, X., Cai, X., Zhang, W., Xu, W., Li, D., Ding, J., Hu, Y., Wang, D., Huang, A., Yin, Y., Wang, D.(2013) PLoS One 8: e68647-e68647
- PubMed: 23874703
- DOI: https://doi.org/10.1371/journal.pone.0068647
- Primary Citation of Related Structures:
4FF5 - PubMed Abstract:
Pathogenic bacteria produce a wide variety of virulence factors that are considered to be potential antibiotic targets. In this study, we report the crystal structure of a novel S. pneumoniae virulence factor, GHIP, which is a streptococcus-specific glycosyl hydrolase. This novel structure exhibits an α/β-barrel fold that slightly differs from other characterized hydrolases. The GHIP active site, located at the negatively charged groove in the barrel, is very similar to the active site in known peptidoglycan hydrolases. Functionally, GHIP exhibited weak enzymatic activity to hydrolyze the PNP-(GlcNAc)5 peptidoglycan by the general acid/base catalytic mechanism. Animal experiments demonstrated a marked attenuation of S. pneumoniae-mediated virulence in mice infected by ΔGHIP-deficient strains, suggesting that GHIP functions as a novel S. pneumoniae virulence factor. Furthermore, GHIP participates in allowing S. pneumoniae to colonize the nasopharynx and invade host epithelial cells. Taken together, these findings suggest that GHIP can potentially serve as an antibiotic target to effectively treat streptococcus-mediated infection.
Organizational Affiliation:
Department of Laboratory Medicine, Chongqing Medical University, Chongqing, People's Republic of China.