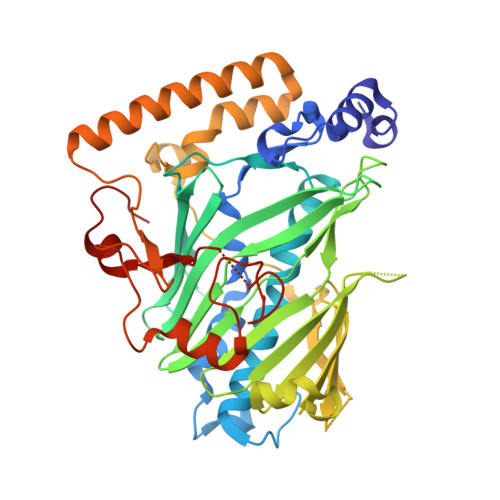
Synthesis and structure of 16,22-diketocholesterol bound to oxysterol-binding protein Osh4.
Koag, M.C., Cheun, Y., Kou, Y., Ouzon-Shubeita, H., Min, K., Monzingo, A.F., Lee, S.(2013) Steroids 78: 938-944
- PubMed: 23756172
- DOI: https://doi.org/10.1016/j.steroids.2013.05.016
- Primary Citation of Related Structures:
4F4B, 4FES - PubMed Abstract:
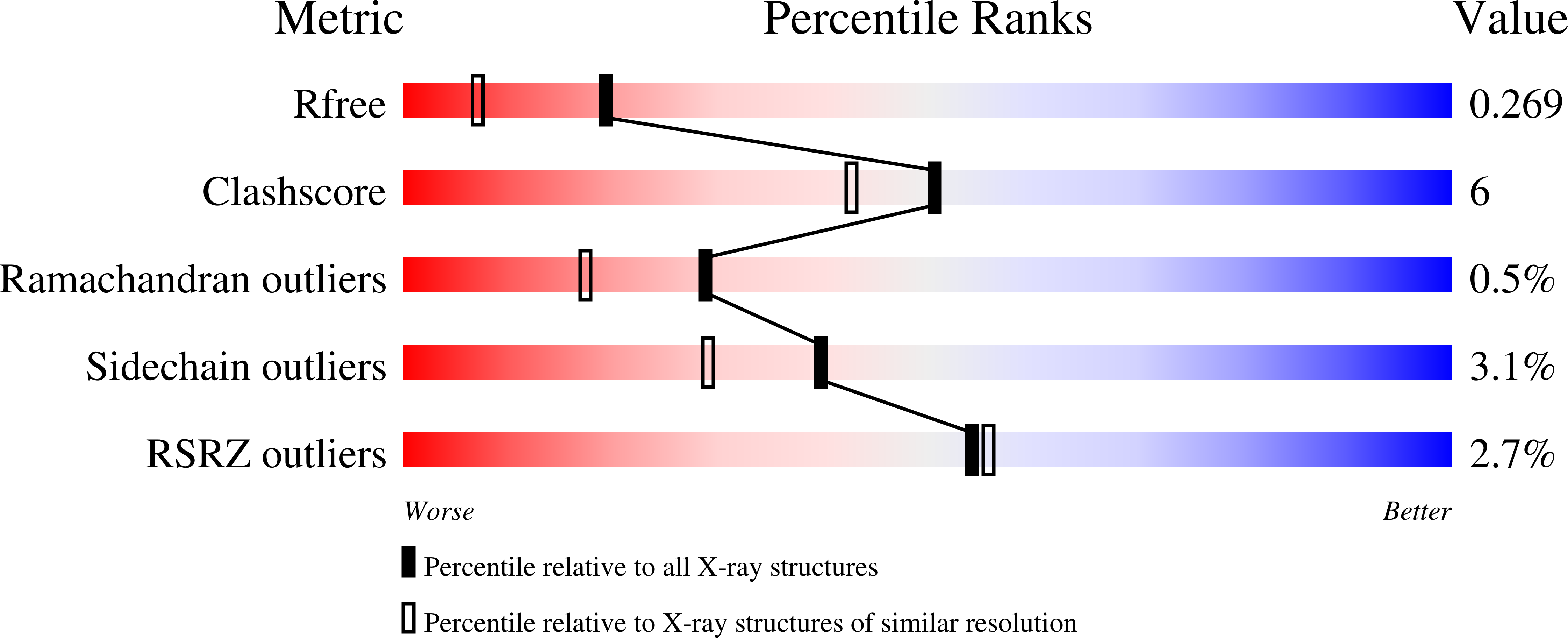
We have synthesized 16,22-diketocholesterol, a novel ligand for oxysterol-binding protein Osh4, and determined X-ray structure of the diketocholesterol in complex with Osh4. The X-ray structure shows that α7 helix of Osh4 assumes open conformation while the rest of Osh4, closed conformation, implying this diketocholesterol-bound Osh4 structure may represent a structural intermediate between the two conformations.
Organizational Affiliation:
Division of Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, Austin, TX 78712, USA.