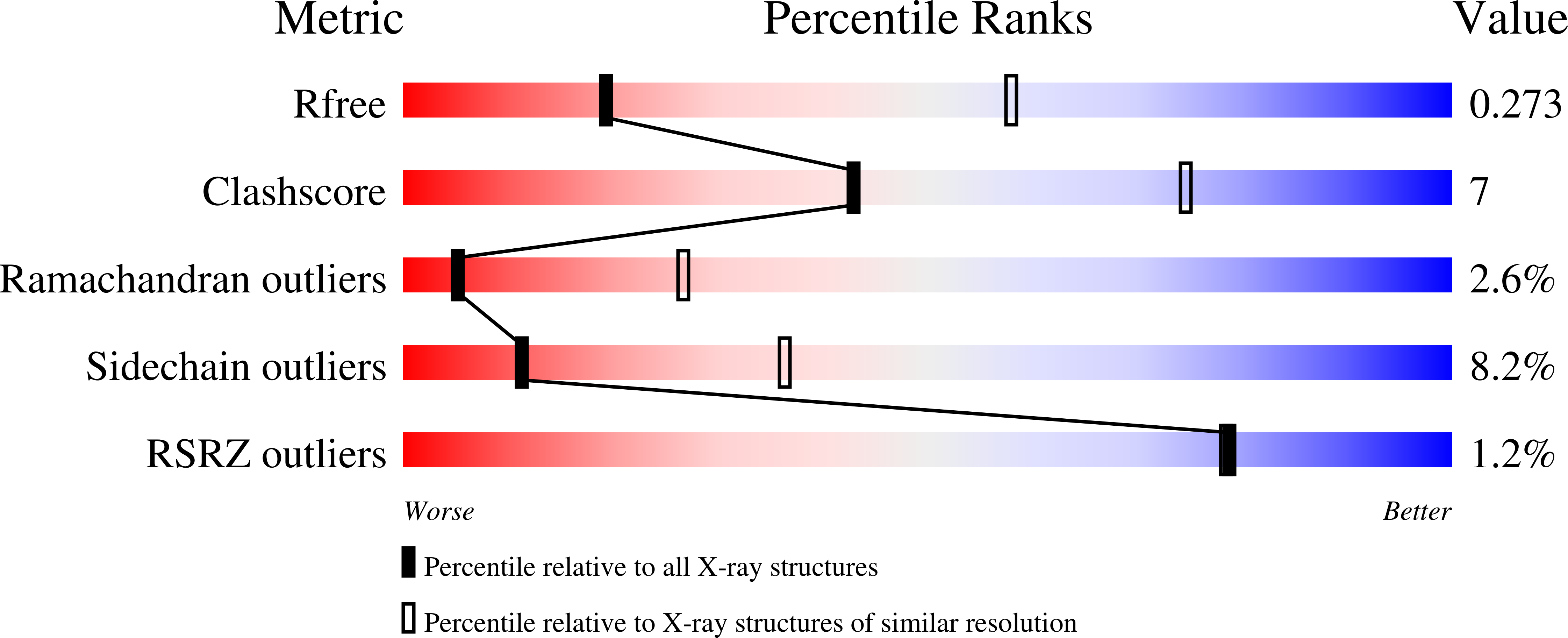
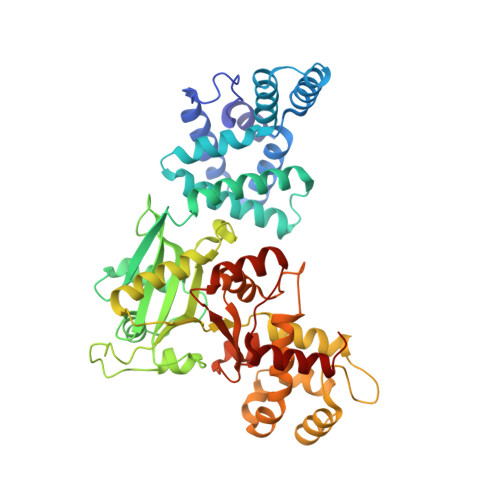
Structural Asymmetry and Disulfide Bridges among Subunits Modulate the Activity of Human Malonyl-CoA Decarboxylase.
Aparicio, D., Perez-Luque, R., Carpena, X., Diaz, M., Ferrer, J.C., Loewen, P.C., Fita, I.(2013) J Biol Chem 288: 11907-11919
- PubMed: 23482565
- DOI: https://doi.org/10.1074/jbc.M112.443846
- Primary Citation of Related Structures:
4F0X - PubMed Abstract:
Decarboxylation of malonyl-CoA to acetyl-CoA by malonyl-CoA decarboxylase (MCD; EC 4.1.1.9) is an essential facet in the regulation of fatty acid metabolism. The structure of human peroxisomal MCD reveals a molecular tetramer that is best described as a dimer of structural heterodimers, in which the two subunits present markedly different conformations. This molecular organization is consistent with half-of-the-sites reactivity. Each subunit has an all-helix N-terminal domain and a catalytic C-terminal domain with an acetyltransferase fold (GNAT superfamily). Intersubunit disulfide bridges, Cys-206-Cys-206 and Cys-243-Cys-243, can link the four subunits of the tetramer, imparting positive cooperativity to the catalytic process. The combination of a half-of-the-sites mechanism within each structural heterodimer and positive cooperativity in the tetramer produces a complex regulatory picture that is further complicated by the multiple intracellular locations of the enzyme. Transport into the peroxisome has been investigated by docking human MCD onto the peroxisomal import protein peroxin 5, which revealed interactions that extend beyond the C-terminal targeting motif.
Organizational Affiliation:
Institut de Biologia Molecular de Barcelona, Consejo Superior de Investigaciones Científicas, 08028 Barcelona, Spain.