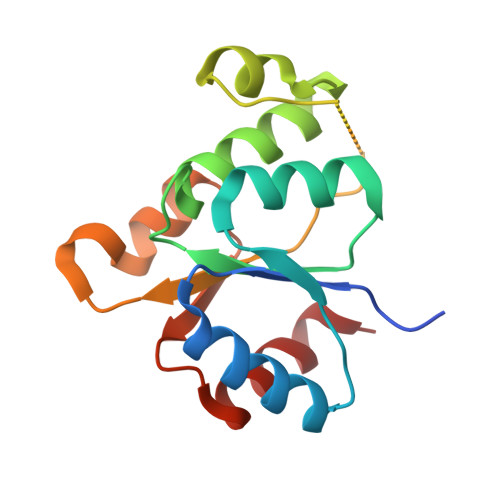
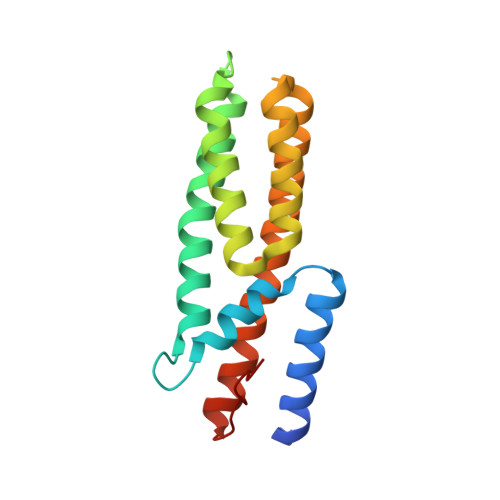
Structure-Function Analysis of Arabidopsis thaliana Histidine Kinase AHK5 Bound to Its Cognate Phosphotransfer Protein AHP1.
Bauer, J., Reiss, K., Veerabagu, M., Heunemann, M., Harter, K., Stehle, T.(2013) Mol Plant 6: 959-970
- PubMed: 23132142
- DOI: https://doi.org/10.1093/mp/sss126
- Primary Citation of Related Structures:
4EUK - PubMed Abstract:
The multi-step phosphorelay (MSP) system defines a key signal transduction pathway in plants and many eukaryotes. In this system, external stimuli first lead to the activation of a histidine kinase, followed by transfer of a phosphoryl group from the receiver domain of the kinase (HK(RD)) to downstream, cytosolic phosphotransfer proteins (HPs). In order to establish the determinants of specificity for this signaling relay system, we have solved the first crystal structure of a plant HK(RD), AHK5(RD), in complex with one of its cognate HPs, AHP1. AHP1 binds AHK5(RD) via a prominent hydrogen bond docking ridge and a hydrophobic patch. These features are conserved among all AHP proteins, but differ significantly from other structurally characterized prokaryotic and eukaryotic HPs. Surface plasmon resonance experiments show that AHK5(RD) binds to AHP1-3 with similar, micromolar affinity, consistent with the transient nature of this signaling complex. Our correlation of structural and functional data provide the first insight, at the atomic level as well as with quantitative affinity data, into the molecular recognition events governing the MSP in plants.
Organizational Affiliation:
Interfaculty Institute of Biochemistry, University of Tübingen, D-72076 Tübingen, Germany.