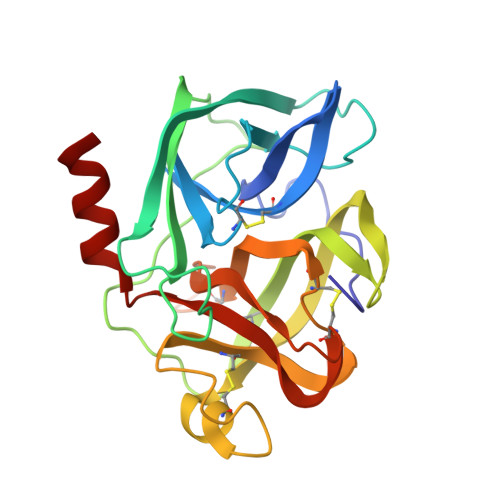
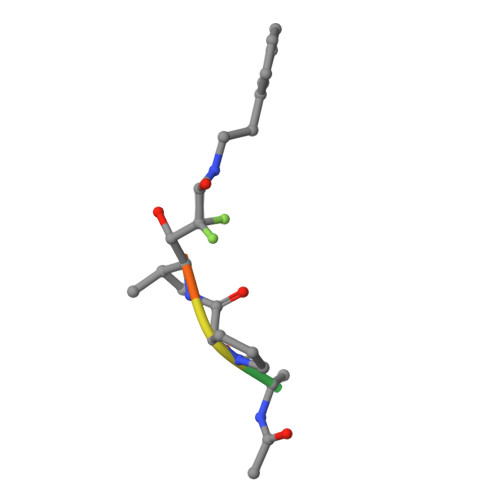
Crystal Structure of the Covalent Complex Formed by a Peptidyl Alpha,Alpha-Difluoro-Beta-Keto Amide with Porcine Pancreatic Elastase at 1.78-Angstroms Resolution
Takahashi, L.H., Radhakrishnan, R., Rosenfieldjunior, R.E., Meyerjunior, E.F., Trainor, D.A.(1989) J Am Chem Soc 111: 3368