Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP
Ghosh, A., Bhoshle, R., Toro, R., Gizzi, A., Hillerich, B., Seidel, R., Almo, S.C.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative oxidoreductase | 255 | Sinorhizobium meliloti 1021 | Mutation(s): 0 Gene Names: R02322, SMc01571 EC: 1 | 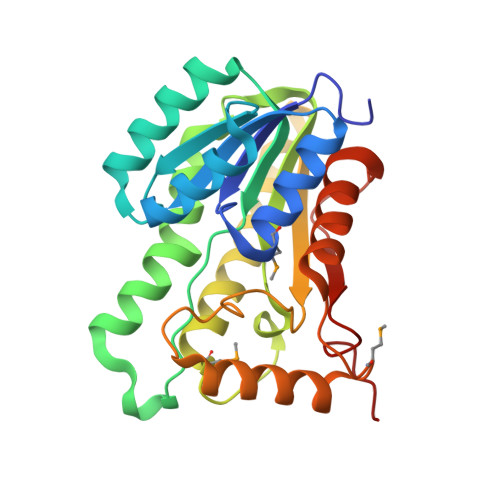 | |
UniProt | |||||
Find proteins for Q92N93 (Rhizobium meliloti (strain 1021)) Explore Q92N93 Go to UniProtKB: Q92N93 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q92N93 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAP Query on NAP | E [auth A], F [auth B], H [auth C], J [auth D] | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE C21 H28 N7 O17 P3 XJLXINKUBYWONI-NNYOXOHSSA-N |  | ||
| GOL Query on GOL | G [auth B], I [auth C] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C, D | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.96 | α = 66.5 |
| b = 66.74 | β = 78.3 |
| c = 70.54 | γ = 62.96 |
| Software Name | Purpose |
|---|---|
| CBASS | data collection |
| SOLVE | phasing |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| SCALEPACK | data scaling |