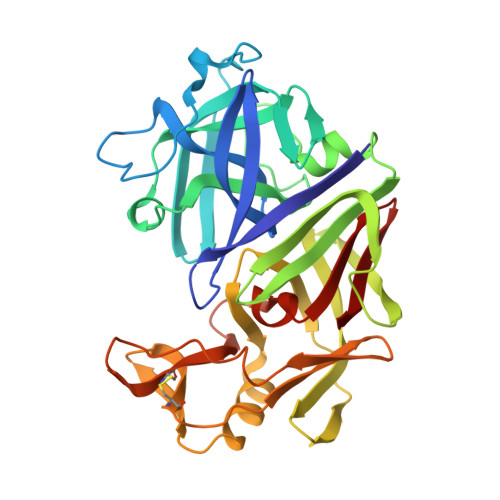
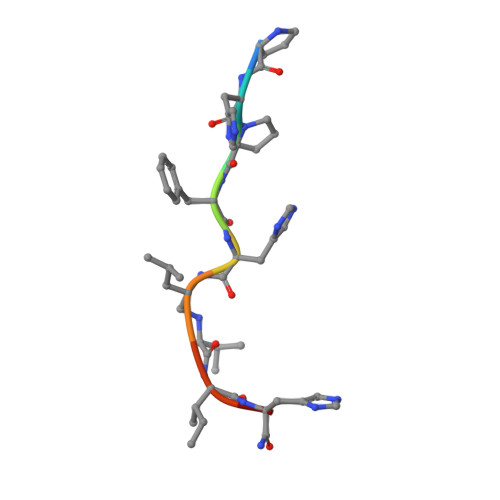
High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Foundling, S.I., Cooper, J., Watson, F.E., Cleasby, A., Pearl, L.H., Sibanda, B.L., Hemmings, A., Wood, S.P., Blundell, T.L., Valler, M.J., Norey, C.G., Kay, J., Boger, J., Dunn, B.M., Leckieparallel, B.J., Jone, D.M., Atrash, B., Hallett, A., Szelke, M.(1987) Nature 327: 349-352
- PubMed: 3295561
- DOI: https://doi.org/10.1038/327349a0
- Primary Citation of Related Structures:
4ER4 - PubMed Abstract:
Inhibitors of the conversion of angiotensinogen to the vasoconstrictor angiotensin II have considerable value as antihypertensive agents. For example, captopril and enalapril are clinically useful as inhibitors of angiotensin-converting enzyme. This has encouraged intense activity in the development of inhibitors of kidney renin, which is a very specific aspartic proteinase catalysing the first and rate limiting step in the conversion of angiotensinogen to angiotensin II. The most effective inhibitors such as H-142 and L-363,564 have used non-hydrolysable analogues of the proposed transition state, and partial sequences of angiotensinogen (Table 1). H-142 is effective in lowering blood pressure in humans but has no significant effect on other aspartic proteinases such as pepsin in the human body (Table 1). At present there are no crystal structures available for human or mouse renins although three-dimensional models demonstrate close structural similarity to other spartic proteinases. We have therefore determined by X-ray analysis the three-dimensional structures of H-142 and L-363,564 complexed with the aspartic proteinase endothiapepsin, which binds these inhibitors with affinities not greatly different from those measured against human renin (Table 1). The structures of these complexes and of that between endothiapepsin and the general aspartic proteinase inhibitor, H-256 (Table 1) define the common hydrogen bonding schemes that allow subtle differences in side-chain orientations and in the positions of the transition state analogues with respect to the active-site aspartates.