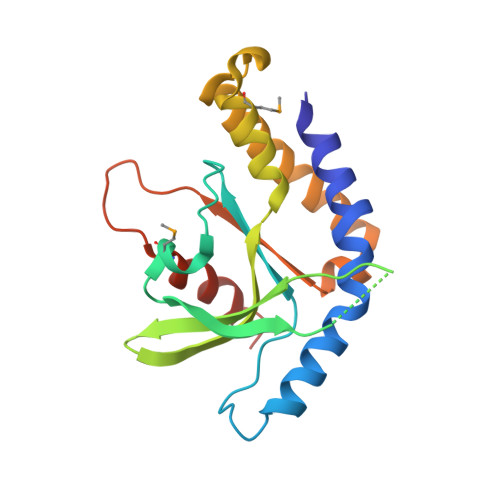
Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Shu, C., Yi, G., Watts, T., Kao, C.C., Li, P.(2012) Nat Struct Mol Biol 19: 722-724
- PubMed: 22728658
- DOI: https://doi.org/10.1038/nsmb.2331
- Primary Citation of Related Structures:
4EMT, 4EMU - PubMed Abstract:
STING (stimulator of interferon genes) is an innate immune sensor of cyclic dinucleotides that regulates the induction of type I interferons. STING's C-terminal domain forms a V-shaped dimer and binds a cyclic diguanylate monophosphate (c-di-GMP) at the dimer interface by both direct and solvent-mediated hydrogen bonds. Guanines of c-di-GMP stack against the phenolic rings of a conserved tyrosine, and mutations at the c-di-GMP binding surface reduce nucleotide binding and affect signaling.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas, USA.