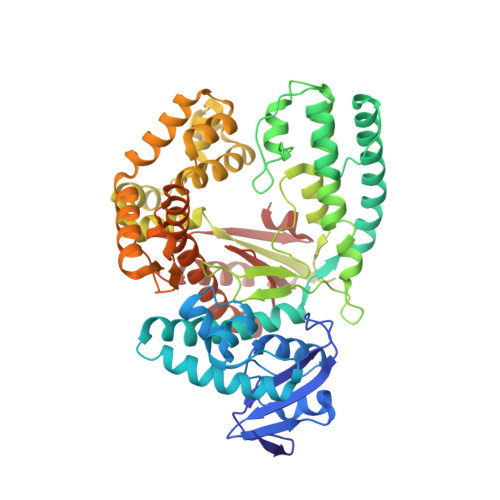
Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site.
Obeid, S., Busskamp, H., Welte, W., Diederichs, K., Marx, A.(2012) Chem Commun (Camb) 48: 8320-8322
- PubMed: 22766607
- DOI: https://doi.org/10.1039/c2cc34181f
- Primary Citation of Related Structures:
4ELT, 4ELU - PubMed Abstract:
Modified nucleotides play a paramount role in many cutting-edge biomolecular techniques. The present structural study highlights the plasticity and flexibility of the active site of a DNA polymerase while incorporating non-polar "Click-able" nucleotide analogs and emphasizes new insights into rational design guidelines for modified nucleotides.
Organizational Affiliation:
Department of Chemistry and Konstanz Research School Chemical Biology, University of Konstanz, Universitätsstrasse 10, 78457 Konstanz, Germany.