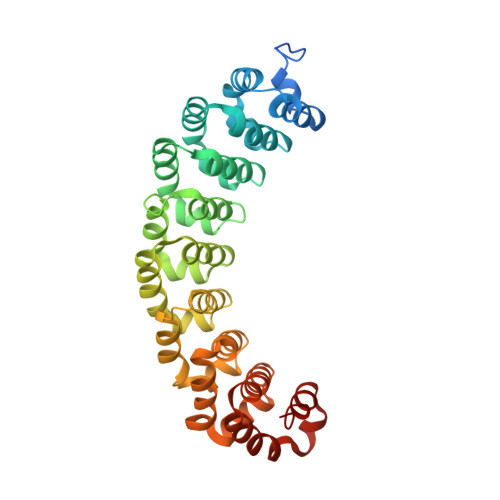
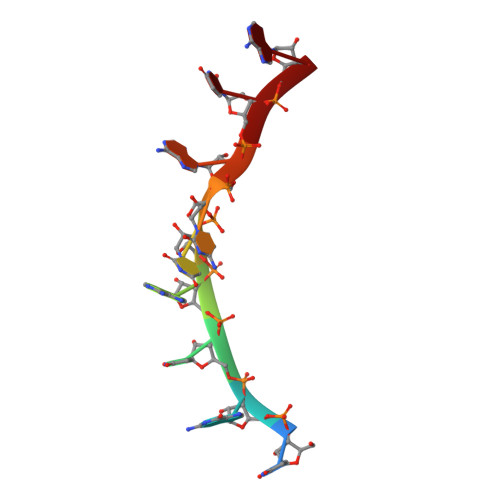
Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site.
Valley, C.T., Porter, D.F., Qiu, C., Campbell, Z.T., Hall, T.M., Wickens, M.(2012) Proc Natl Acad Sci U S A 109: 6054-6059
- PubMed: 22467831
- DOI: https://doi.org/10.1073/pnas.1200521109
- Primary Citation of Related Structures:
4DZS - PubMed Abstract:
mRNA control hinges on the specificity and affinity of proteins for their RNA binding sites. Regulatory proteins must bind their own sites and reject even closely related noncognate sites. In the PUF [Pumilio and fem-3 binding factor (FBF)] family of RNA binding proteins, individual proteins discriminate differences in the length and sequence of binding sites, allowing each PUF to bind a distinct battery of mRNAs. Here, we show that despite these differences, the pattern of RNA interactions is conserved among PUF proteins: the two ends of the PUF protein make critical contacts with the two ends of the RNA sites. Despite this conserved "two-handed" pattern of recognition, the RNA sequence is flexible. Among the binding sites of yeast Puf4p, RNA sequence dictates the pattern in which RNA bases are flipped away from the binding surface of the protein. Small differences in RNA sequence allow new modes of control, recruiting Puf5p in addition to Puf4p to a single site. This embedded information adds a new layer of biological meaning to the connections between RNA targets and PUF proteins.
Organizational Affiliation:
Graduate Program in Cellular and Molecular Biology, University of Wisconsin, Madison, WI 53706, USA.