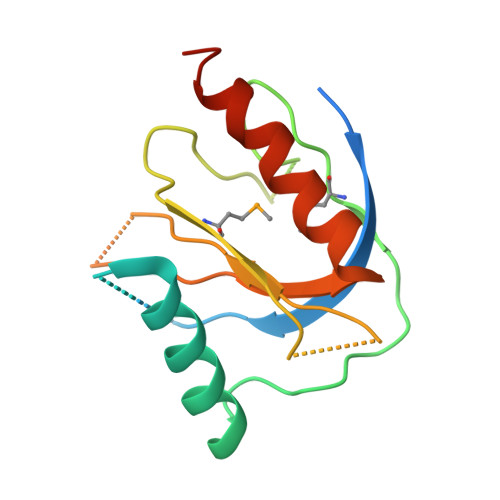
Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Liu, W., Draheim, K.M., Zhang, R., Calderwood, D.A., Boggon, T.J.(2013) Mol Cell 49: 719-729
- PubMed: 23317506
- DOI: https://doi.org/10.1016/j.molcel.2012.12.005
- Primary Citation of Related Structures:
4DX8, 4DX9 - PubMed Abstract:
KRIT1 (Krev/Rap1 Interaction Trapped-1) mutations are observed in ∼40% of autosomal-dominant cerebral cavernous malformations (CCMs), a disease occurring in up to 0.5% of the population. We show that KRIT1 functions as a switch for β1 integrin activation by antagonizing ICAP1 (Integrin Cytoplasmic Associated Protein-1)-mediated modulation of "inside-out" activation. We present cocrystal structures of KRIT1 with ICAP1 and ICAP1 with integrin β1 cytoplasmic tail to 2.54 and 3.0 Å resolution (the resolutions at which I/σI = 2 are 2.75 and 3.0 Å, respectively). We find that KRIT1 binds ICAP1 by a bidentate surface, that KRIT1 directly competes with integrin β1 to bind ICAP1, and that KRIT1 antagonizes ICAP1-modulated integrin activation using this site. We also find that KRIT1 contains an N-terminal Nudix domain, in a region previously designated as unstructured. We therefore provide insights to integrin regulation and CCM-associated KRIT1 function.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06520, USA.