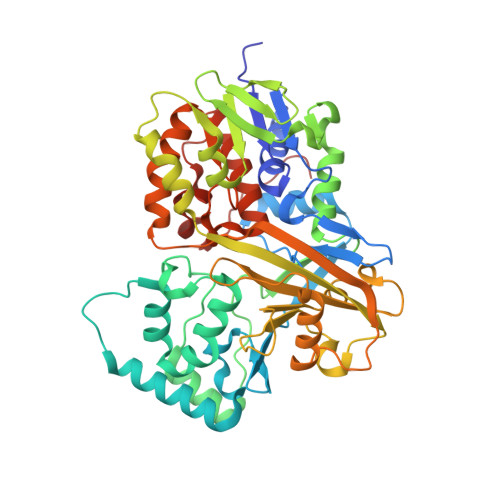
Crystal Structures of Trypanosoma cruzi UDP-Galactopyranose Mutase Implicate Flexibility of the Histidine Loop in Enzyme Activation.
Dhatwalia, R., Singh, H., Oppenheimer, M., Sobrado, P., Tanner, J.J.(2012) Biochemistry 51: 4968-4979
- PubMed: 22646091
- DOI: https://doi.org/10.1021/bi300498c
- Primary Citation of Related Structures:
4DSG, 4DSH - PubMed Abstract:
Chagas disease is a neglected tropical disease caused by the protozoan parasite Trypanosoma cruzi. Here we report crystal structures of the galactofuranose biosynthetic enzyme UDP-galactopyranose mutase (UGM) from T. cruzi, which are the first structures of this enzyme from a protozoan parasite. UGM is an attractive target for drug design because galactofuranose is absent in humans but is an essential component of key glycoproteins and glycolipids in trypanosomatids. Analysis of the enzyme-UDP noncovalent interactions and sequence alignments suggests that substrate recognition is exquisitely conserved among eukaryotic UGMs and distinct from that of bacterial UGMs. This observation has implications for inhibitor design. Activation of the enzyme via reduction of the FAD induces profound conformational changes, including a 2.3 Å movement of the histidine loop (Gly60-Gly61-His62), rotation and protonation of the imidazole of His62, and cooperative movement of residues located on the si face of the FAD. Interestingly, these changes are substantially different from those described for Aspergillus fumigatus UGM, which is 45% identical to T. cruzi UGM. The importance of Gly61 and His62 for enzymatic activity was studied with the site-directed mutant enzymes G61A, G61P, and H62A. These mutations lower the catalytic efficiency by factors of 10-50, primarily by decreasing k(cat). Considered together, the structural, kinetic, and sequence data suggest that the middle Gly of the histidine loop imparts flexibility that is essential for activation of eukaryotic UGMs. Our results provide new information about UGM biochemistry and suggest a unified strategy for designing inhibitors of UGMs from the eukaryotic pathogens.
Organizational Affiliation:
Department of Chemistry, University of Missouri-Columbia, Columbia, Missouri 65211, USA.