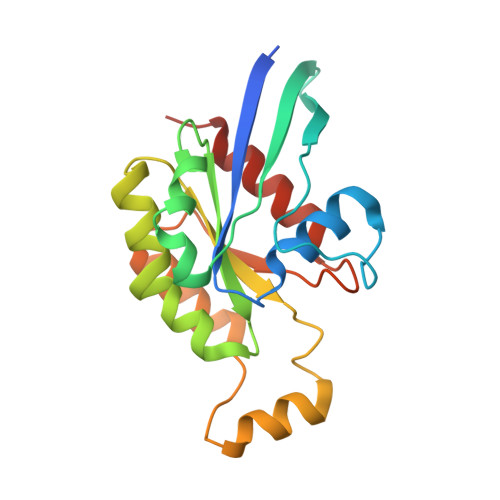
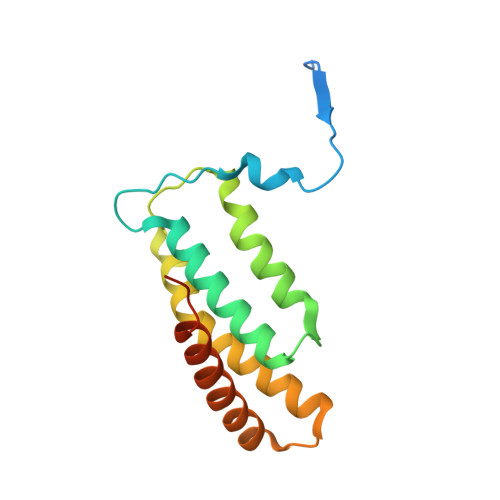
Structure of Salmonella Effector Protein SopB N-terminal Domain in Complex with Host Rho GTPase Cdc42.
Burkinshaw, B.J., Prehna, G., Worrall, L.J., Strynadka, N.C.(2012) J Biol Chem 287: 13348-13355
- PubMed: 22362774
- DOI: https://doi.org/10.1074/jbc.M111.331330
- Primary Citation of Related Structures:
4DID - PubMed Abstract:
SopB is a type III secreted Salmonella effector protein with phosphoinositide phosphatase activity and a distinct GTPase binding domain. The latter interacts with host Cdc42, an essential Rho GTPase that regulates critical events in eukaryotic cytoskeleton organization and membrane trafficking. Structural and biochemical analysis of the SopB GTPase binding domain in complex with Cdc42 shows for the first time that SopB structurally and functionally mimics a host guanine nucleotide dissociation inhibitor (GDI) by contacting key residues in the regulatory switch regions of Cdc42 and slowing Cdc42 nucleotide exchange.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology and Center for Blood Research, University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada.