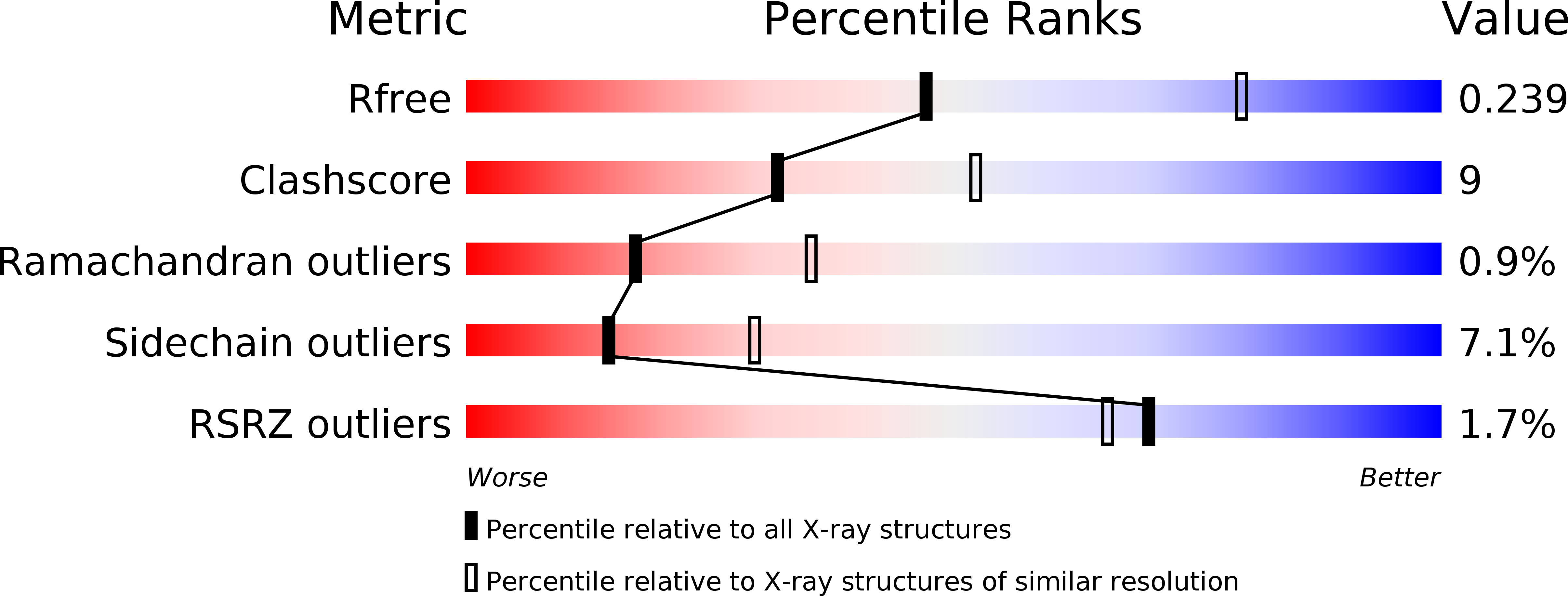
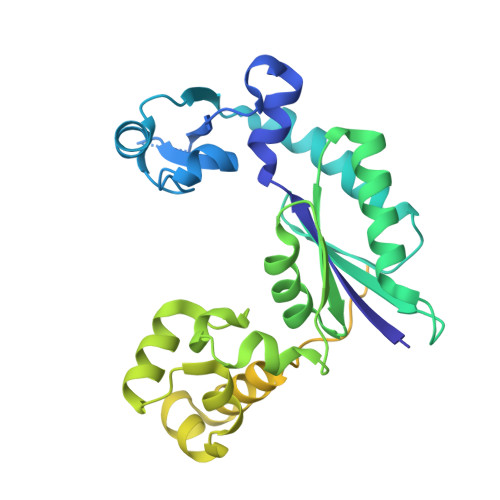
The PAD region in the mycobacterial DinB homologue MsPolIV exhibits positional heterogeneity.
Sharma, A., Subramanian, V., Nair, D.T.(2012) Acta Crystallogr D Biol Crystallogr 68: 960-967
- PubMed: 22868761
- DOI: https://doi.org/10.1107/S0907444912017623
- Primary Citation of Related Structures:
4DEZ - PubMed Abstract:
Y-family DNA polymerases (dPols) have evolved to carry out translesion bypass to rescue stalled replication; prokaryotic members of this family also participate in the phenomenon of adaptive mutagenesis to relieve selection pressure imposed by a maladapted environment. In this study, the first structure of a member of this family from a prokaryote has been determined. The structure of MsPolIV, a Y-family dPol from Mycobacterium smegmatis, shows the presence of the characteristic finger, palm and thumb domains. Surprisingly, the electron-density map of the intact protein does not show density for the PAD region that is unique to members of this family. Analysis of the packing of the molecules in the crystals showed the existence of large solvent-filled voids in which the PAD region could be located in multiple conformations. In line with this observation, analytical gel-filtration and dynamic light-scattering studies showed that MsPolIV undergoes significant compaction upon DNA binding. The PAD region is known to insert into the major groove of the substrate DNA and to play a major role in shaping the active site. Comparison with structures of other Y-family dPols shows that in the absence of tertiary contacts between the PAD domain and the other domains this region has the freedom to adopt multiple orientations. This structural attribute of the PAD will allow these enzymes to accommodate the alterations in the width of the DNA double helix that are necessary to achieve translesion bypass and adaptive mutagenesis and will also allow regulation of their activity to prevent adventitious error-prone DNA synthesis.
Organizational Affiliation:
National Centre for Biological Sciences (NCBS-TIFR), UAS-GKVK Campus, Bellary Road, Bangalore 560 065, India.