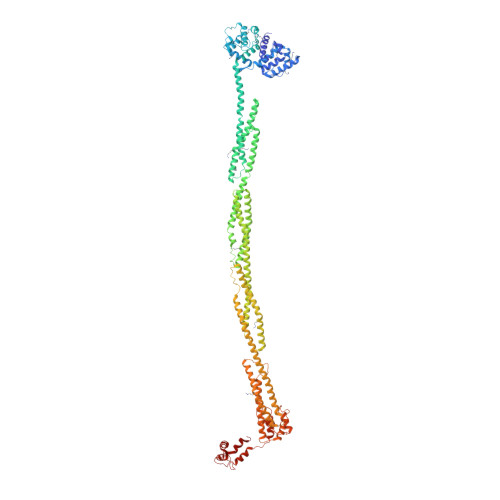The Structure and Regulation of Human Muscle Alpha-Actinin
Ribeiro Jr, E.A., Pinotsis, N., Ghisleni, A., Salmazo, A., Konarev, P.V., Kostan, J., Sjoeblom, B., Schreiner, C., Polyansky, A.A., Gkougkoulia, E., Holt, M.R., Aachmann, F.L., Zagrovic, B., Bordignon, E., Pirker, K.F., Svergun, D.I., Gautel, M., Djinovic-Carugo, K.(2014) Cell 159: 1447
- PubMed: 25433700
- DOI: https://doi.org/10.1016/j.cell.2014.10.056
- Primary Citation of Related Structures:
4D1E - PubMed Abstract:
The spectrin superfamily of proteins plays key roles in assembling the actin cytoskeleton in various cell types, crosslinks actin filaments, and acts as scaffolds for the assembly of large protein complexes involved in structural integrity and mechanosensation, as well as cell signaling. α-actinins in particular are the major actin crosslinkers in muscle Z-disks, focal adhesions, and actin stress fibers. We report a complete high-resolution structure of the 200 kDa α-actinin-2 dimer from striated muscle and explore its functional implications on the biochemical and cellular level. The structure provides insight into the phosphoinositide-based mechanism controlling its interaction with sarcomeric proteins such as titin, lays a foundation for studying the impact of pathogenic mutations at molecular resolution, and is likely to be broadly relevant for the regulation of spectrin-like proteins.
Organizational Affiliation:
Department of Structural and Computational Biology, Max F. Perutz Laboratories, University of Vienna, Campus Vienna Biocenter 5, 1030 Vienna, Austria.