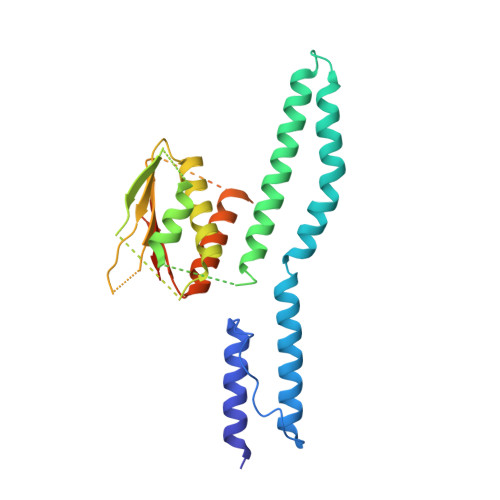
Crystallographic Snapshot of the Escherichia Coli Envz Histidine Kinase in an Active Conformation.
Ferris, H.U., Coles, M., Lupas, A.N., Hartmann, M.D.(2014) J Struct Biol 186: 376
- PubMed: 24681325
- DOI: https://doi.org/10.1016/j.jsb.2014.03.014
- Primary Citation of Related Structures:
4CTI - PubMed Abstract:
Sensor histidine kinases are important sensors of the extracellular environment and relay signals via conformational changes that trigger autophosphorylation of the kinase and subsequent phosphorylation of a response regulator. The exact mechanism and the regulation of this protein family are a matter of ongoing investigation. Here we present a crystal structure of a functional chimeric protein encompassing the entire catalytic part of the Escherichia coli EnvZ histidine kinase, fused to the HAMP domain of the Archaeoglobus fulgidus Af1503 receptor. The construct is thus equivalent to the full cytosolic part of EnvZ. The structure shows a putatively active conformation of the catalytic domain and gives insight into how this conformation could be brought about in response to sensory input. Our analysis suggests a sequential flip-flop autokinase mechanism.
Organizational Affiliation:
Department of Protein Evolution, Max-Planck-Institute for Developmental Biology, D-72076 Tübingen, Germany.