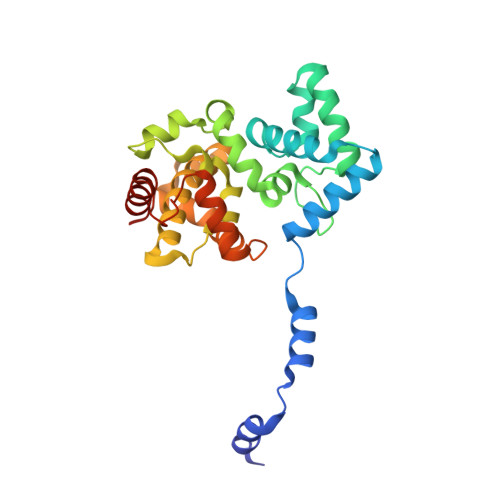
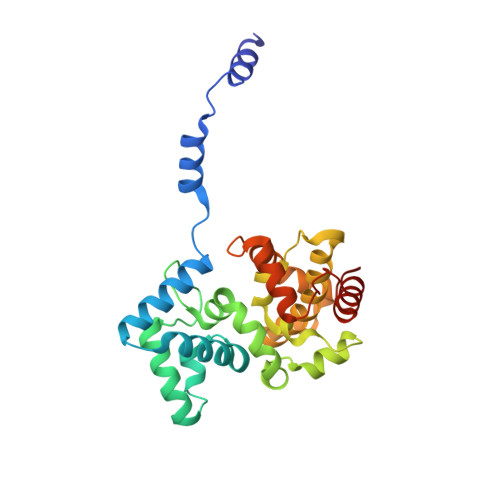
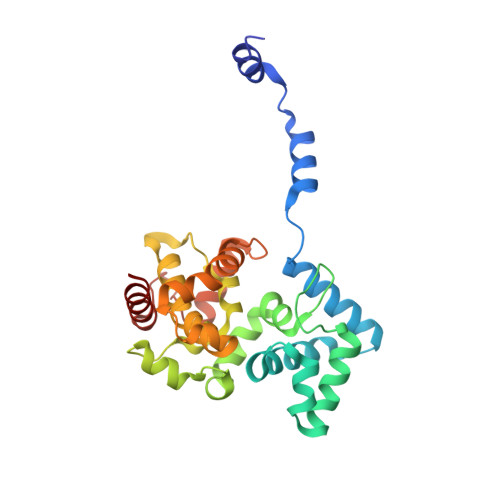
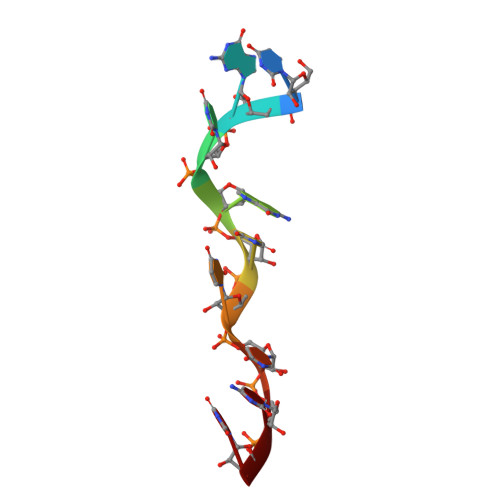Structural Insights Into RNA Encapsidation and Helical Assembly of the Toscana Virus Nucleoprotein.
Olal, D., Dick, A., Woods, V.L., Liu, T., Li, S., Devignot, S., Weber, F., Saphire, E.O., Daumke, O.(2014) Nucleic Acids Res 42: 6025
- PubMed: 24688060
- DOI: https://doi.org/10.1093/nar/gku229
- Primary Citation of Related Structures:
4CSF, 4CSG - PubMed Abstract:
Toscana virus is an emerging bunyavirus in Mediterranean Europe where it accounts for 80% of pediatric meningitis cases during the summer. The negative-strand ribonucleic acid (RNA) genome of the virus is wrapped around the virally encoded nucleoprotein N to form the ribonucleoprotein complex (RNP). We determined crystal structures of hexameric N alone (apo) and in complex with a nonameric single-stranded RNA. RNA is sequestered in a sequence-independent fashion in a deep groove inside the hexamer. At the junction between two adjacent copies of Ns, RNA binding induced an inter-subunit rotation, which opened the RNA-binding tunnel and created a new assembly interface at the outside of the hexamer. Based on these findings, we suggest a structural model for how binding of RNA to N promotes the formation of helical RNPs, which are a characteristic hallmark of many negative-strand RNA viruses.
Organizational Affiliation:
Max Delbrück Center for Molecular Medicine, Crystallography, Robert-Rössle-Strasse 10, 13125 Berlin, Germany Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA 92037, USA oliver.daumke@mdc-berlin.de.