Structural Insights Into the Inhibition of Wnt Signaling by Cancer Antigen 5T4/Wnt-Activated Inhibitory Factor 1.
Zhao, Y., Malinauskas, T., Harlos, K., Jones, E.Y.(2014) Structure 22: 612
- PubMed: 24582434
- DOI: https://doi.org/10.1016/j.str.2014.01.009
- Primary Citation of Related Structures:
4CNC, 4CNM - PubMed Abstract:
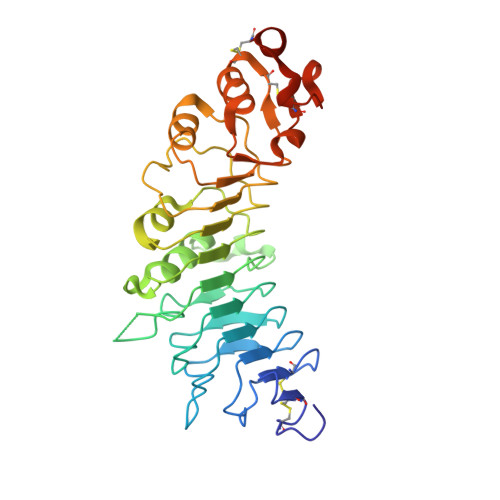
The tumor antigen 5T4/WAIF1 (Wnt-activated inhibitory factor 1; also known as Trophoblast glycoprotein TPBG) is a cell surface protein targeted in multiple cancer immunotherapy clinical trials. Recently, it has been shown that 5T4/WAIF1 inhibits Wnt/β-catenin signaling, a signaling system central to many developmental and pathological processes. Wnt/β-catenin signaling is controlled by multiple inhibitors and activators. Here, we report crystal structures for the extracellular domain of 5T4/WAIF1 at 1.8 Å resolution. They reveal a highly glycosylated, rigid core, comprising eight leucine-rich repeats (LRRs), which serves as a platform to present evolutionarily conserved surface residues in the N-terminal LRR1. Structural and cell-based analyses, coupled with previously reported in vivo data, suggest that Tyr325 plus the LRR1 surface centered on a second exposed aromatic residue, Phe97, are essential for inhibition of Wnt/β-catenin signaling. These results provide a structural basis for the development of 5T4/WAIF1-targeted therapies that preserve or block 5T4/WAIF1-mediated inhibition of Wnt/β-catenin signaling.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, UK.