Structure and Regulatory Targets of Sco3201, a Highly Promiscuous Tetr-Like Regulator of Streptomyces Coelicolor M145.
Xu, D., Waack, P., Zhang, Q., Werten, S., Hinrichs, W., Virolle, M.(2014) Biochem Biophys Res Commun 450: 513
- PubMed: 24928397
- DOI: https://doi.org/10.1016/j.bbrc.2014.06.003
- Primary Citation of Related Structures:
4CGR - PubMed Abstract:
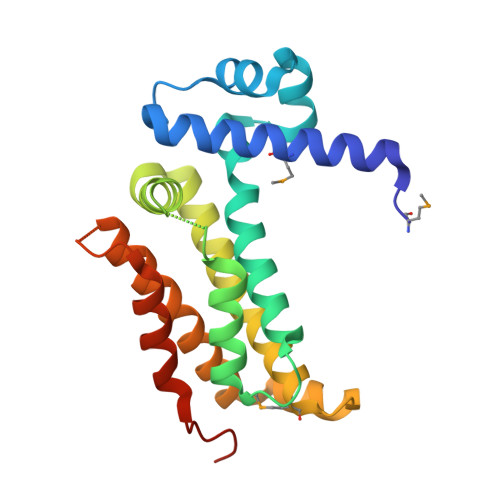
SCO3201, a regulator of the TetR family, is a strong repressor of both morphological differentiation and antibiotic production when overexpressed in Streptomyces coelicolor. Here, we report the identification of 14 novel putative regulatory targets of this regulator using in vitro formaldehyde cross-linking. Direct binding of purified His6-SCO3201 was demonstrated for the promoter regions of 5 regulators (SCO1716, SCO1950, SCO3367, SCO4009 and SCO5046), a putative histidine phosphatase (SCO1809), an acetyltransferase (SCO0988) and the polyketide synthase RedX (SCO5878), using EMSA. Reverse transcriptional analysis demonstrated that the expression of the transcriptional regulators SCO1950, SCO4009, SCO5046, as well as of SCO0988 and RedX was down regulated, upon SCO3201 overexpression, whereas the expression of SCO1809 and SCO3367 was up regulated. A consensus binding motif was derived via alignment of the promoter regions of the genes negatively regulated. The positions of the predicted operator sites were consistent with a direct repressive effect of SCO3201 on 5 out of 7 of these promoters. Furthermore, the 2.1Å crystal structure of SCO3201 was solved, which provides a possible explanation for the high promiscuity of this regulator that might account for its dramatic effect on the differentiation process of S. coelicolor.
Organizational Affiliation:
Department of Ecology, Institute of Hydrobiology, School of Life Science and Technology, Jinan University, Guangzhou 510632, PR China. Electronic address: xudelin@live.com.