The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
Edlich-Muth, C., Artero, J., Callow, P., Przewloka, M.R., Watson, A.A., Zhang, W., Glover, D.M., Debski, J., Dadlez, M., Round, A.R., Trevor Forsyth, V., Laue, E.D.(2015) J Mol Biol 427: 1949
- PubMed: 25813344
- DOI: https://doi.org/10.1016/j.jmb.2015.03.010
- Primary Citation of Related Structures:
4CA9 - PubMed Abstract:
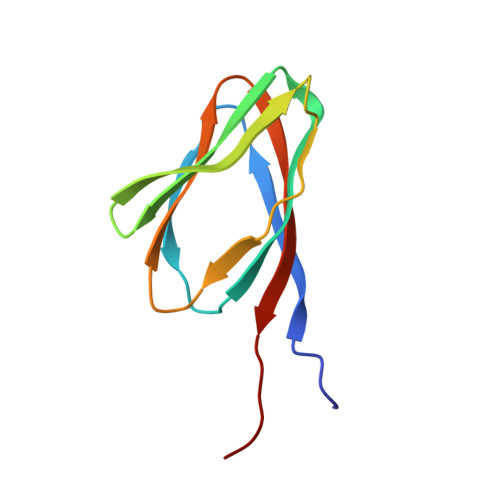
Nucleoplasmin is a histone chaperone that consists of a pentameric N-terminal domain and an unstructured C-terminal tail. The pentameric core domain, a doughnut-like structure with a central pore, is only found in the nucleoplasmin family. Here, we report the first structure of a nucleoplasmin-like domain (NPL) from the unrelated Drosophila protein, FKBP39, and we present evidence that this protein associates with chromatin. Furthermore, we show that two other chromatin proteins, Arabidopsis thaliana histone deacetylase type 2 (HD2) and Saccharomyces cerevisiae Fpr4, share the NPL fold and form pentamers, or a dimer of pentamers in the case of HD2. Thus, we propose a new family of proteins that share the pentameric nucleoplasmin-like NPL domain and are found in protists, fungi, plants and animals.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, CB2 1GA Cambridge, United Kingdom.