Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
Deiss, S., Hernandez Alvarez, B., Bar, K., Ewers, C.P., Coles, M., Albrecht, R., Hartmann, M.D.(2014) J Struct Biol 186: 380
- PubMed: 24486584
- DOI: https://doi.org/10.1016/j.jsb.2014.01.013
- Primary Citation of Related Structures:
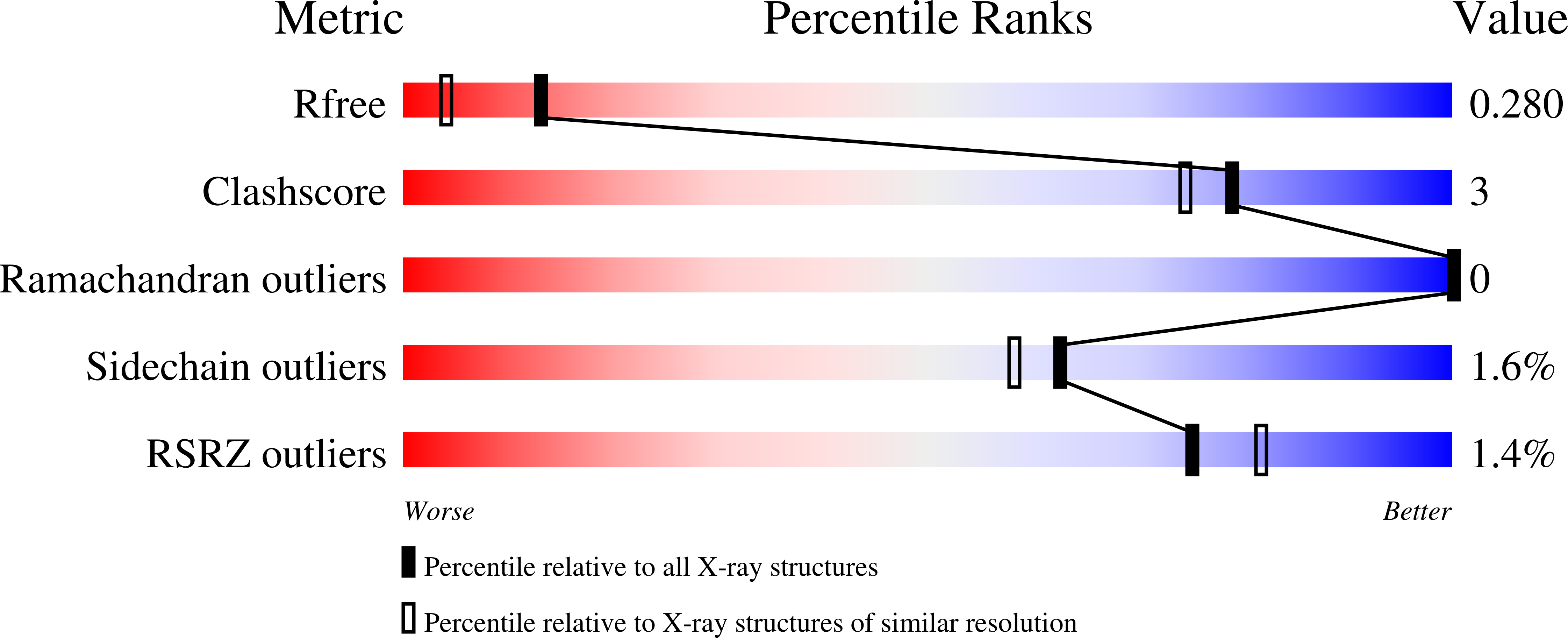
4C46 - PubMed Abstract:
This work presents a protein structure that has been designed purely for aesthetic reasons, symbolizing decades of coiled-coil research and praising its most fundamental model system, the GCN4 leucine zipper. The GCN4 leucine zipper is a highly stable coiled coil which can be tuned to adopt different oligomeric states via mutation of its core residues. For these reasons it is used in structural studies as a stabilizing fusion adaptor. On the occasion of the 50th birthday of Andrei N. Lupas, we used it to create the first personalized protein structure: we fused the sequence ANDREI-N-LVPAS in heptad register to trimeric GCN4 adaptors and determined its structure by X-ray crystallography. The structure demonstrates the robustness and versatility of GCN4 as a fusion adaptor. We learn how proline can be accommodated in trimeric coiled coils, and put the structure into the context of the other GCN4-fusion structures known to date.
Organizational Affiliation:
Department of Protein Evolution, Max Planck Institute for Developmental Biology, Spemannstraße 35, D-72076 Tübingen, Germany.