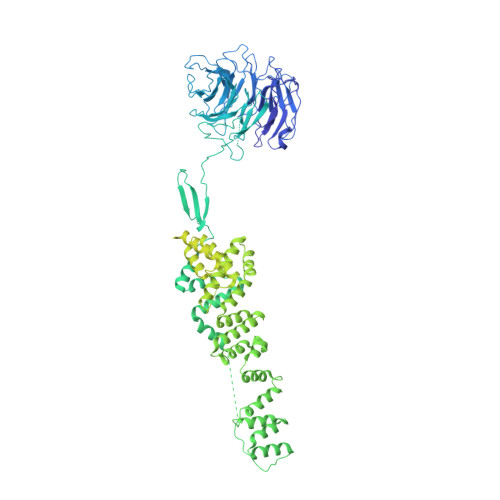
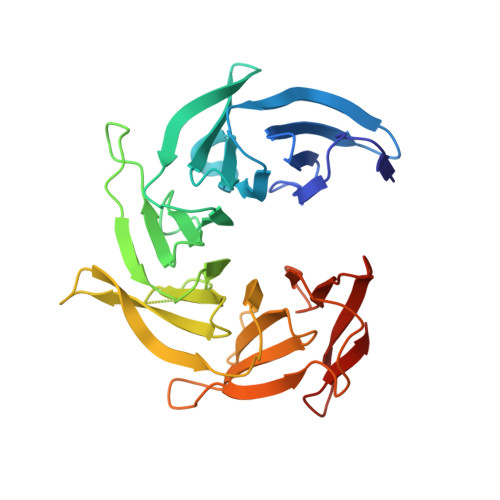
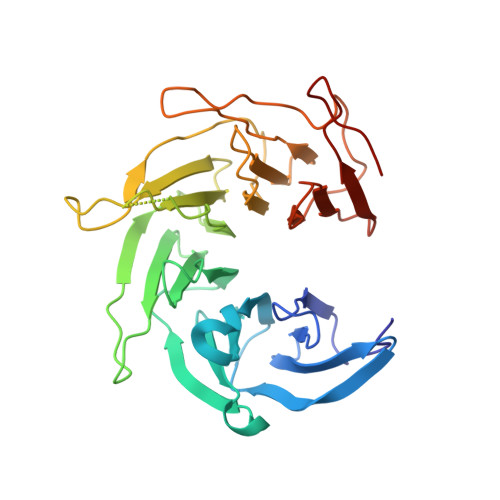
The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Zanetti, G., Prinz, S., Daum, S., Meister, A., Schekman, R., Bacia, K., Briggs, J.A.G.(2013) Elife 2: 00951
- PubMed: 24062940
- DOI: https://doi.org/10.7554/eLife.00951
- Primary Citation of Related Structures:
4BZI, 4BZJ, 4BZK - PubMed Abstract:
Coat protein complex II (COPII) mediates formation of the membrane vesicles that export newly synthesised proteins from the endoplasmic reticulum. The inner COPII proteins bind to cargo and membrane, linking them to the outer COPII components that form a cage around the vesicle. Regulated flexibility in coat architecture is essential for transport of a variety of differently sized cargoes, but structural data on the assembled coat has not been available. We have used cryo-electron tomography and subtomogram averaging to determine the structure of the complete, membrane-assembled COPII coat. We describe a novel arrangement of the outer coat and find that the inner coat can assemble into regular lattices. The data reveal how coat subunits interact with one another and with the membrane, suggesting how coordinated assembly of inner and outer coats can mediate and regulate packaging of vesicles ranging from small spheres to large tubular carriers. DOI:http://dx.doi.org/10.7554/eLife.00951.001.
Organizational Affiliation:
Department of Molecular and Cell Biology , University of California, Berkeley , Berkeley , United States.