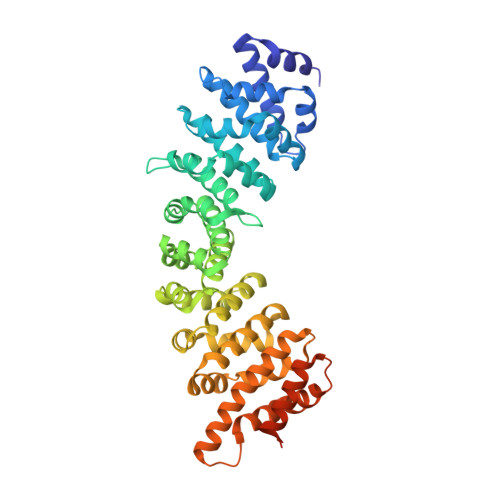
Structural Basis of Interaction of Bipartite Nuclear Localization Signal from Agrobacterium Vird2 with Rice Importin-Alpha
Chang, C.-W., Williams, S.J., Counago, R.M., Kobe, B.(2014) Mol Plant 7: 1061
- PubMed: 24503158
- DOI: https://doi.org/10.1093/mp/ssu014
- Primary Citation of Related Structures:
4BPL, 4BQK
Organizational Affiliation:
School of Chemistry and Molecular Biosciences, Institute for Molecular Bioscience, and Australian Infectious Diseases Research Centre, University of Queensland, Brisbane, Qld 4072, Australia.