The Crystal Structure of the Cell Division Amidase Amic Reveals the Fold of the Amin Domain, a New Peptidoglycan Binding Domain.
Rocaboy, M., Herman, R., Sauvage, E., Remaut, H., Moonens, K., Terrak, M., Charlier, P., Kerff, F.(2013) Mol Microbiol 90: 267
- PubMed: 23927005
- DOI: https://doi.org/10.1111/mmi.12361
- Primary Citation of Related Structures:
4BIN - PubMed Abstract:
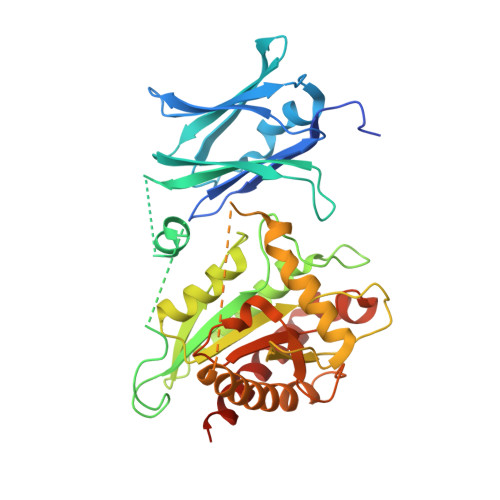
Binary fission is the ultimate step of the prokaryotic cell cycle. In Gram-negative bacteria like Escherichia coli, this step implies the invagination of three biological layers (cytoplasmic membrane, peptidoglycan and outer membrane), biosynthesis of the new poles and eventually, daughter cells separation. The latter requires the coordinated action of the N-acetylmuramyl-L-alanine amidases AmiA/B/C and their LytM activators EnvC and NlpD to cleave the septal peptidoglycan. We present here the 2.5 Å crystal structure of AmiC which includes the first report of an AMIN domain structure, a β-sandwich of two symmetrical four-stranded β-sheets exposing highly conserved motifs on the two outer faces. We show that this N-terminal domain, involved in the localization of AmiC at the division site, is a new peptidoglycan-binding domain. The C-terminal catalytic domain shows an auto-inhibitory alpha helix obstructing the active site. AmiC lacking this helix exhibits by itself an activity comparable to that of the wild type AmiC activated by NlpD. We also demonstrate the interaction between AmiC and NlpD by microscale thermophoresis and confirm the importance of the active site blocking alpha helix in the regulation of the amidase activity.
Organizational Affiliation:
Centre d'Ingénierie des Protéines, University of Liège, Liège, Belgium.