1.2 A X-Ray Structure of the Renal Potassium Channel Kv1.3 T1 Domain.
Kremer, W., Weyand, M., Winklmeier, A., Schreier, C., Kalbitzer, H.R.(2013) Protein J 32: 533
- PubMed: 24114469
- DOI: https://doi.org/10.1007/s10930-013-9513-2
- Primary Citation of Related Structures:
4BGC - PubMed Abstract:
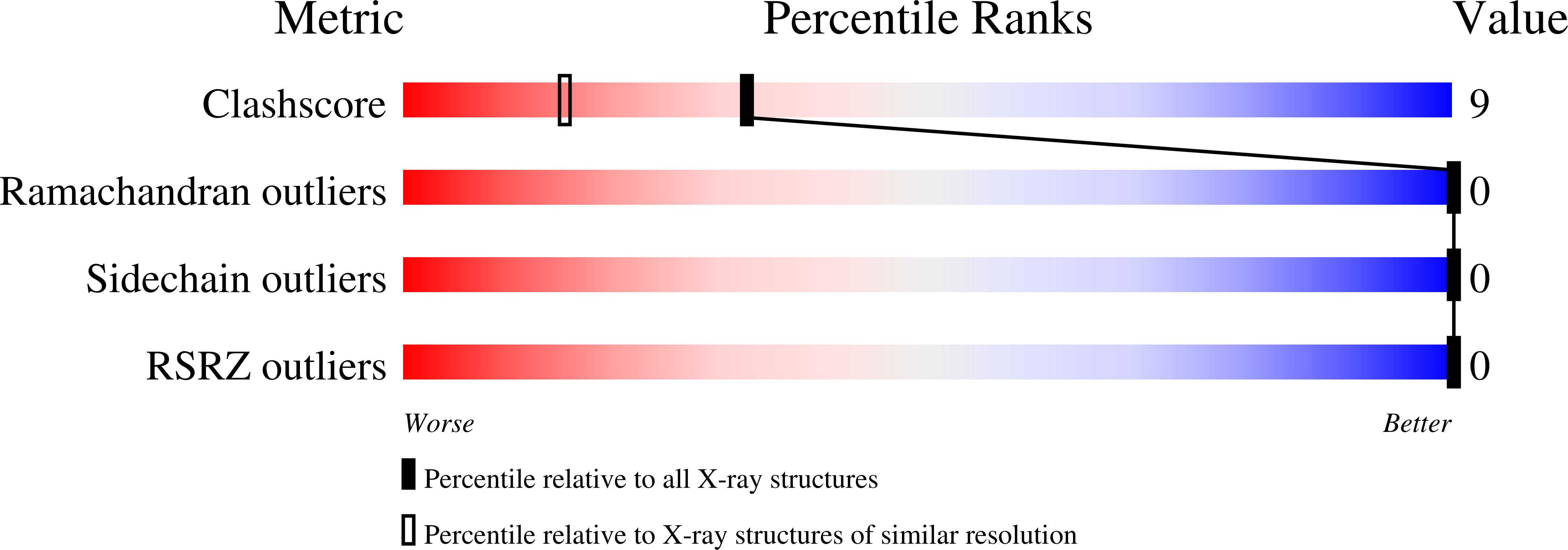
Here we present the structure of the T1 domain derived from the voltage-dependent potassium channel K(v)1.3 of Homo sapiens sapiens at 1.2 Å resolution crystallized under near-physiological conditions. The crystals were grown without precipitant in 150 mM KP(i), pH 6.25. The crystals show I4 symmetry typical of the natural occurring tetrameric assembly of the single subunits. The obtained structural model is based on the highest resolution currently achieved for tetramerization domains of voltage-gated potassium channels. We identified an identical fold of the monomer but inside the tetramer the single monomers show a significant rotation which leads to a different orientation of the tetramer compared to other known structures. Such a rotational movement inside the tetrameric assembly might influence the gating properties of the channel. In addition we see two distinct side chain configurations for amino acids located in the top layer proximal to the membrane (Tyr109, Arg116, Ser129, Glu140, Met142, Arg146), and amino acids in the bottom layer of the T1-domain distal from the membrane (Val55, Ile56, Leu77, Arg86). The relative populations of these two states are ranging from 50:50 for Val55, Tyr109, Arg116, Ser129, Glu140, 60:40 for Met142, 65:35 for Arg86, 70:30 for Arg146, and 80:20 for Ile56 and Leu77. The data suggest that in solution these amino acids are involved in an equilibrium of conformational states that may be coupled to the functional states of the whole potassium channel.
Organizational Affiliation:
Department of Biophysics and Physical Biochemistry, Centre of Magnetic Resonance in Chemistry and Biomedicine, University of Regensburg, Universitätsstraße 31, 93053, Regensburg, Germany, werner.kremer@ur.de.