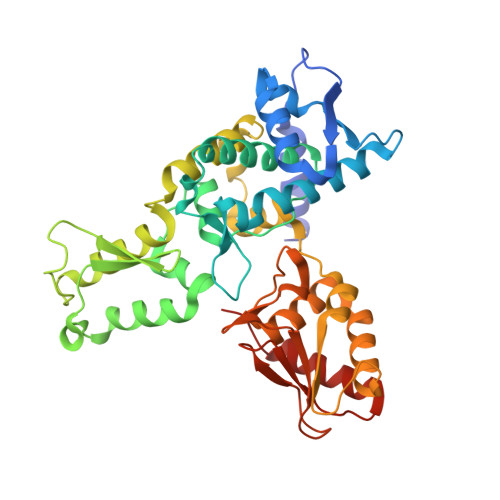
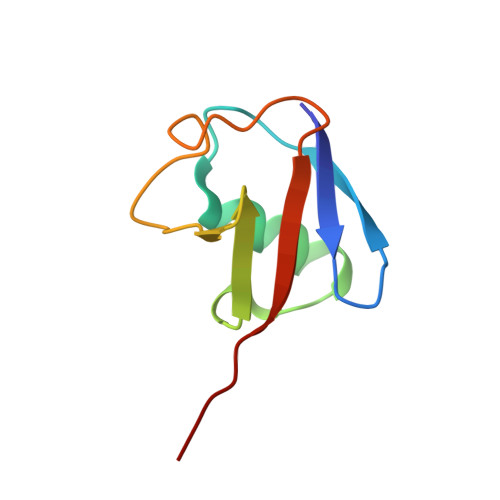
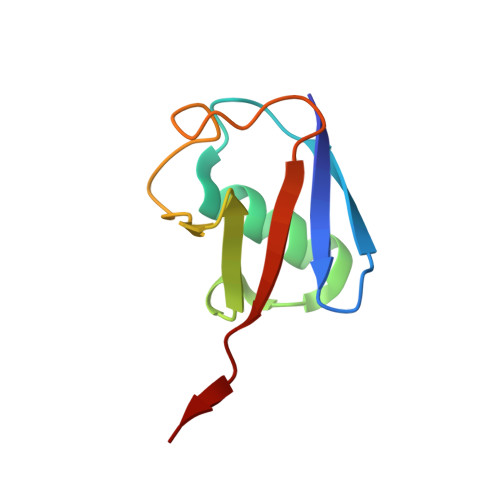
Structure of a Ubiquitin-Loaded Hect Ligase Reveals the Molecular Basis for Catalytic Priming
Maspero, E., Valentini, E., Mari, S., Cecatiello, V., Soffientini, P., Pasqualato, S., Polo, S.(2013) Nat Struct Mol Biol 20: 696
- PubMed: 23644597
- DOI: https://doi.org/10.1038/nsmb.2566
- Primary Citation of Related Structures:
4BBN, 4BE8 - PubMed Abstract:
Homologous to E6-AP C terminus (HECT) E3 ligases recognize and directly catalyze ligation of ubiquitin (Ub) to their substrates. Molecular details of this process remain unknown. We report the first structure, to our knowledge, of a Ub-loaded E3, the human neural precursor cell-expressed developmentally downregulated protein 4 (Nedd4). The HECT(Nedd4)~Ub transitory intermediate provides a structural basis for the proposed sequential addition mechanism. The donor Ub, transferred from the E2, is bound to the Nedd4 C lobe with its C-terminal tail locked in an extended conformation, primed for catalysis. We provide evidence that the Nedd4-family members are Lys63-specific enzymes whose catalysis is mediated by an essential C-terminal acidic residue.
Organizational Affiliation:
Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy.