Brms151-98 and Brms151-84 are Crystal Oligomeric Coiled Coils with Different Oligomerization States, which Behave as Disordered Protein Fragments in Solution.
Spinola-Amilibia, M., Rivera, J., Ortiz-Lombardia, M., Romero, A., Neira, J.L., Bravo, J.(2013) J Mol Biol 425: 2147
- PubMed: 23500495
- DOI: https://doi.org/10.1016/j.jmb.2013.03.005
- Primary Citation of Related Structures:
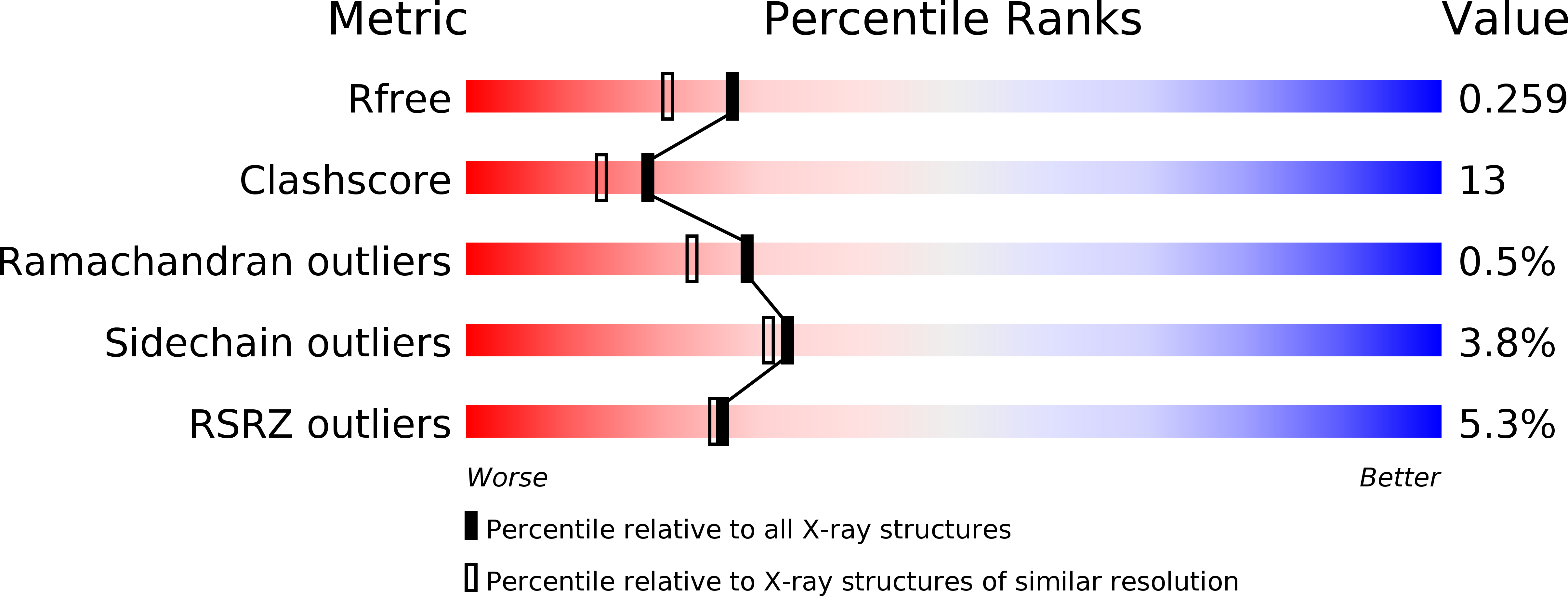
4AUV - PubMed Abstract:
The breast cancer metastasis suppressor 1 (BRMS1) gene suppresses metastasis without affecting the primary tumor growth. Cellular localization of BRMS1 appears to be important for exerting its effects on metastasis inhibition. We recently described a nucleo-cytoplasmic shuttling for BRMS1 and identified a nuclear export signal within the N-terminal coiled coil. The structure of these regions shows an antiparallel coiled coil capable of oligomerizing, which compromises the accessibility to the nuclear export signal consensus residues. We have studied the structural and biophysical features of this region to further understand the contribution of the N-terminal coiled coil to the biological function of BRMS1. We have observed that residues 85 to 98 might be important in defining the oligomerization state of the BRMS1 N-terminal coiled coil. The fragments are mainly disordered in solution, with evidence of residual structure. In addition, we report the presence of a conformational dynamic equilibrium (oligomeric folded species ↔ oligomeric unfolded) in solution in the BRMS1 N-terminal coiled coil that might facilitate the nuclear export of BRMS1 to the cytoplasm.
Organizational Affiliation:
Instituto de Biomedicina de Valencia (IBV-CSIC), C/Jaime Roig 11, 46010 Valencia, Spain.