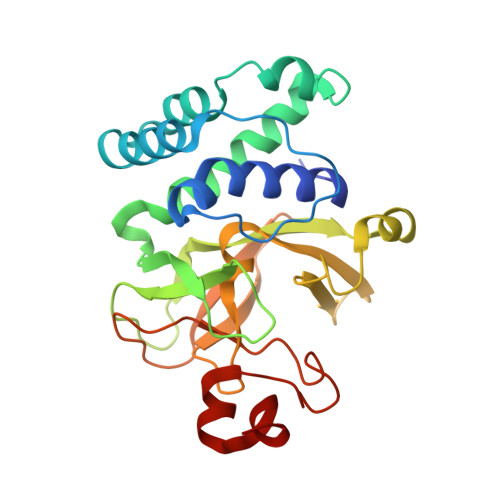
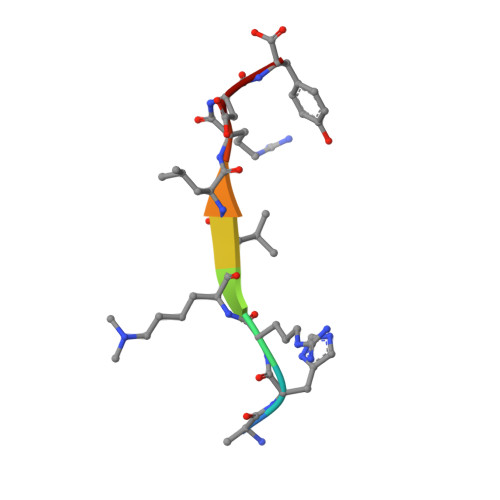
A Novel Route to Product Specificity in the Suv4-20 Family of Histone H4K20 Methyltransferases.
Southall, S.M., Cronin, N.B., Wilson, J.R.(2014) Nucleic Acids Res 42: 661
- PubMed: 24049080
- DOI: https://doi.org/10.1093/nar/gkt776
- Primary Citation of Related Structures:
4AU7, 4BUP - PubMed Abstract:
The delivery of site-specific post-translational modifications to histones generates an epigenetic regulatory network that directs fundamental DNA-mediated processes and governs key stages in development. Methylation of histone H4 lysine-20 has been implicated in DNA repair, transcriptional silencing, genomic stability and regulation of replication. We present the structure of the histone H4K20 methyltransferase Suv4-20h2 in complex with its histone H4 peptide substrate and S-adenosyl methionine cofactor. Analysis of the structure reveals that the Suv4-20h2 active site diverges from the canonical SET domain configuration and generates a high degree of both substrate and product specificity. Together with supporting biochemical data comparing Suv4-20h1 and Suv4-20h2, we demonstrate that the Suv4-20 family enzymes take a previously mono-methylated H4K20 substrate and generate an exclusively di-methylated product. We therefore predict that other enzymes are responsible for the tri-methylation of histone H4K20 that marks silenced heterochromatin.
Organizational Affiliation:
Division of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, London, SW3 6JB, UK.