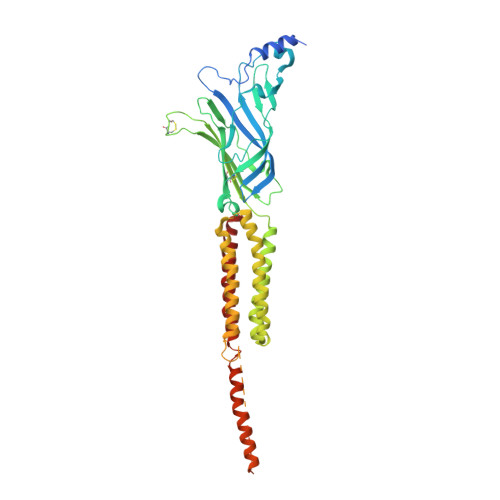
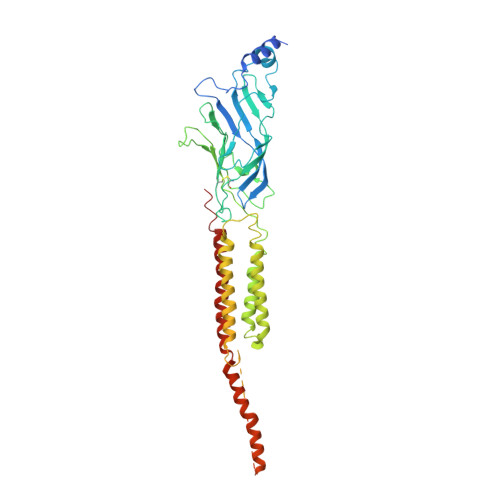
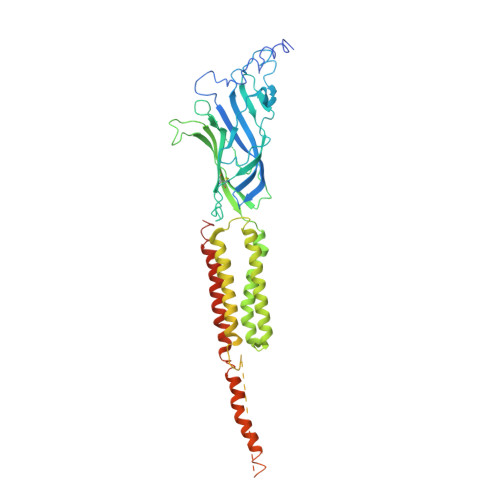
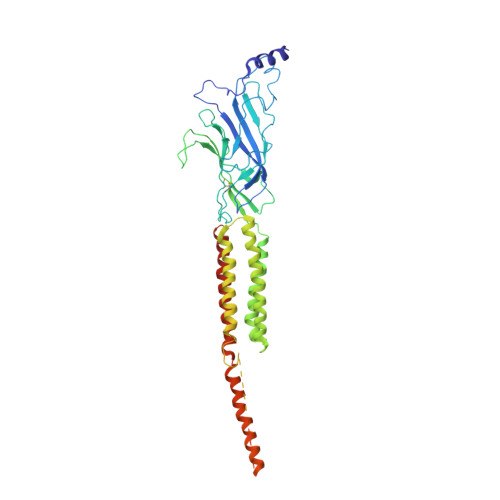
Gating Movement of Acetylcholine Receptor Caught by Plunge-Freezing.
Unwin, N., Fujiyoshi, Y.(2012) J Mol Biol 422: 617
- PubMed: 22841691
- DOI: https://doi.org/10.1016/j.jmb.2012.07.010
- Primary Citation of Related Structures:
4AQ5, 4AQ9 - PubMed Abstract:
The nicotinic acetylcholine (ACh) receptor converts transiently to an open-channel form when activated by ACh released into the synaptic cleft. We describe here the conformational change underlying this event, determined by electron microscopy of ACh-sprayed and freeze-trapped postsynaptic membranes. ACh binding to the α subunits triggers a concerted rearrangement in the ligand-binding domain, involving an ~1-Å outward displacement of the extracellular portion of the β subunit where it interacts with the juxtaposed ends of α-helices shaping the narrow membrane-spanning pore. The β-subunit helices tilt outward to accommodate this displacement, destabilising the arrangement of pore-lining helices, which in the closed channel bend inward symmetrically to form a central hydrophobic gate. Straightening and tangential motion of the pore-lining helices effect channel opening by widening the pore asymmetrically and increasing its polarity in the region of the gate. The pore-lining helices of the α(γ) and δ subunits, by flexing between alternative bent and straight conformations, undergo the greatest movements. This coupled allosteric transition shifts the structure from a tense (closed) state toward a more relaxed (open) state.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK. Electronic address: unwin@mrc-lmb.cam.ac.uk.