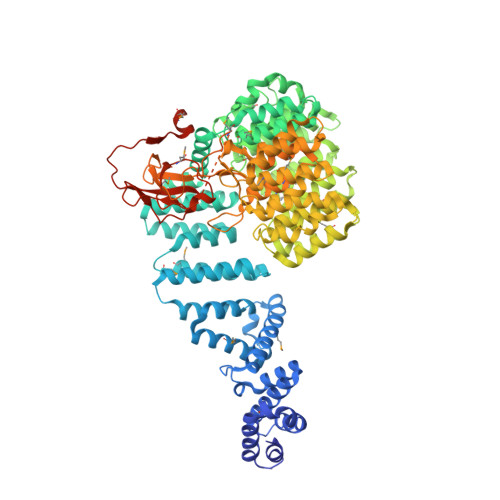
The Structure of the 26S Proteasome Subunit Rpn2 Reveals its Pc Repeat Domain as a Closed Toroid of Two Concentric Alpha-Helical Rings
He, J., Kulkarni, K., Da Fonseca, P.C.A., Krutauz, D., Glickman, M.H., Barford, D., Morris, E.P.(2012) Structure 20: 513
- PubMed: 22405010
- DOI: https://doi.org/10.1016/j.str.2011.12.015
- Primary Citation of Related Structures:
4ADY - PubMed Abstract:
The 26S proteasome proteolyses ubiquitylated proteins and is assembled from a 20S proteolytic core and two 19S regulatory particles (19S-RP). The 19S-RP scaffolding subunits Rpn1 and Rpn2 function to engage ubiquitin receptors. Rpn1 and Rpn2 are characterized by eleven tandem copies of a 35-40 amino acid repeat motif termed the proteasome/cyclosome (PC) repeat. Here, we reveal that the eleven PC repeats of Rpn2 form a closed toroidal structure incorporating two concentric rings of α helices encircling two axial α helices. A rod-like N-terminal domain consisting of 17 stacked α helices and a globular C-terminal domain emerge from one face of the toroid. Rpn13, an ubiquitin receptor, binds to the C-terminal 20 residues of Rpn2. Rpn1 adopts a similar conformation to Rpn2 but differs in the orientation of its rod-like N-terminal domain. These findings have implications for understanding how 19S-RPs recognize, unfold, and deliver ubiquitylated substrates to the 20S core.
Organizational Affiliation:
Division of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, United Kingdom.