An Unexpected Duo: Rubredoxin Binds Nine TPR Motifs to Form LapB, an Essential Regulator of Lipopolysaccharide Synthesis.
Prince, C., Jia, Z.(2015) Structure 23: 1500-1506
- PubMed: 26190574
- DOI: https://doi.org/10.1016/j.str.2015.06.011
- Primary Citation of Related Structures:
4ZLH - PubMed Abstract:
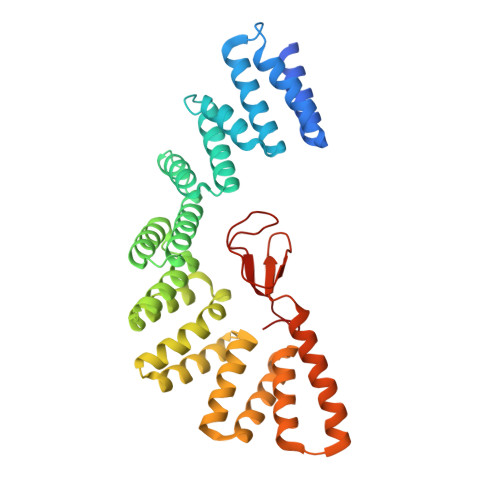
Lipopolysaccharide (LPS) synthesis and export are essential pathways for bacterial growth, proliferation, and virulence. The essential protein LapB from Escherichia coli has recently been identified as a regulator of LPS synthesis. We have determined the crystal structure of LapB (without the N-terminal transmembrane helix) at 2 Å resolution using zinc single-wavelength anomalous diffraction phasing derived from a single bound zinc atom. This structure demonstrates the presence of nine tetratricopeptide repeats (TPR) motifs, including two TPR folds that were not predicted from sequence, and a rubredoxin-type metal binding domain. The rubredoxin domain is bound intimately to the TPR motifs, which has not been previously observed or predicted. Mutations in the rubredoxin/TPR interface inhibit in vivo cell growth, and in vitro studies indicate that these modifications cause local displacement of rubredoxin from its binding site without changing the secondary structure of LapB. LapB is the first reported structure to contain both a rubredoxin domain and TPR motifs.
Organizational Affiliation:
Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON K7L3N6, Canada.