4YY7
The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with avian receptor analog 3'SLNLN
- PDB DOI: https://doi.org/10.2210/pdb4YY7/pdb
- Classification: IMMUNE SYSTEM
- Organism(s): H6N1 subtype
- Expression System: Insect cell expression vector pTIE1
- Mutation(s): No
- Deposited: 2015-03-23 Released: 2016-03-23
- Funding Organization(s): China Ministry of Science and Technology National 973 Project, Intramural Special Grant for Influenza Virus Research from the Chinese Academy of Sciences, Intramural Special Grant for Strategic Priority Research Program of the Chinese Academy of Sciences, National Natural Science Foundation of China, China National Grand S&T Special Project
Experimental Data Snapshot
- Method: X-RAY DIFFRACTION
- Resolution: 2.99 Å
- R-Value Free: 0.226
- R-Value Work: 0.199
- R-Value Observed: 0.200
This is version 2.0 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HA1 | 325 | H6N1 subtype | Mutation(s): 0 | 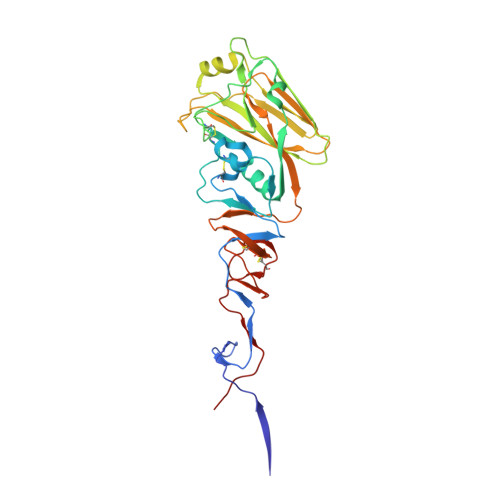 | |
UniProt | |||||
Find proteins for A0A0J9X268 (H6N1 subtype) Explore A0A0J9X268 Go to UniProtKB: A0A0J9X268 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0J9X268 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HA2 | 164 | H6N1 subtype | Mutation(s): 0 | 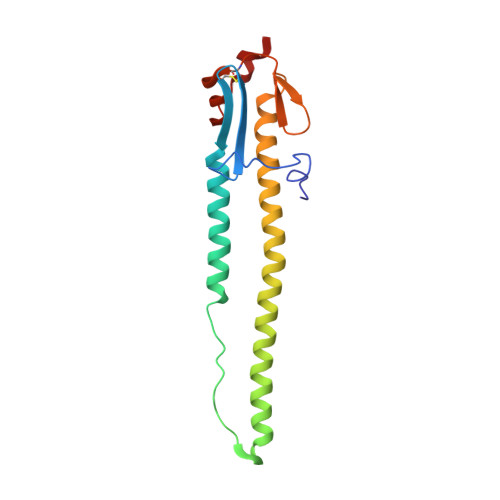 | |
UniProt | |||||
Find proteins for A0A0J9X267 (H6N1 subtype) Explore A0A0J9X267 Go to UniProtKB: A0A0J9X267 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0J9X267 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Small Molecules
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | I [auth A], J [auth B], K [auth C], L [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
Biologically Interesting Molecules (External Reference) 1 Unique
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900067 Query on PRD_900067 | F, H | 3'-sialyl-N-acetyllactosamine | Oligosaccharide / Substrate analog |  | |
Experimental Data & Validation
Experimental Data
- Method: X-RAY DIFFRACTION
- Resolution: 2.99 Å
- R-Value Free: 0.226
- R-Value Work: 0.199
- R-Value Observed: 0.200
- Space Group: P 3
Unit Cell:
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.772 | α = 90 |
| b = 96.772 | β = 90 |
| c = 132.506 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data processing |
| PHASER | phasing |
| Coot | model building |
| PHENIX | refinement |
| SCALA | data reduction |
| SCALEPACK | data scaling |
Entry History & Funding Information
Deposition Data
- Released Date: 2016-03-23 Deposition Author(s): Wang, F., Qi, J., Bi, Y., Zhang, W., Wang, M., Wang, M., Liu, J., Yan, J., Shi, Y., Gao, G.F.
| Funding Organization | Location | Grant Number |
|---|---|---|
| China Ministry of Science and Technology National 973 Project | China | 2011CB504703 |
| Intramural Special Grant for Influenza Virus Research from the Chinese Academy of Sciences | China | KJZD-EW-L09 |
| Intramural Special Grant for Strategic Priority Research Program of the Chinese Academy of Sciences | China | XDB08020100 |
| National Natural Science Foundation of China | China | 31402196 |
| China National Grand S&T Special Project | China | 2014ZX10004002 |
Revision History (Full details and data files)
- Version 1.0: 2016-03-23
Type: Initial release - Version 1.1: 2016-04-20
Changes: Database references - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Atomic model, Data collection, Database references, Derived calculations, Structure summary















