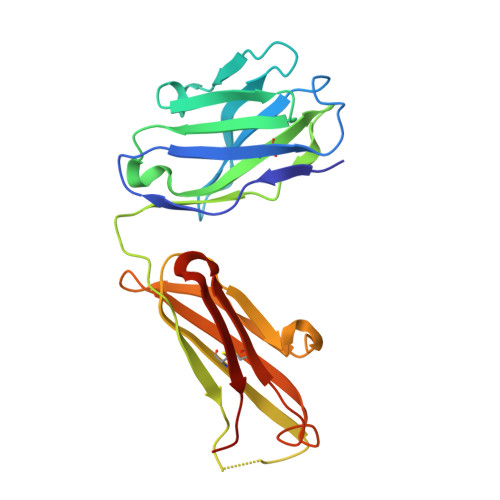
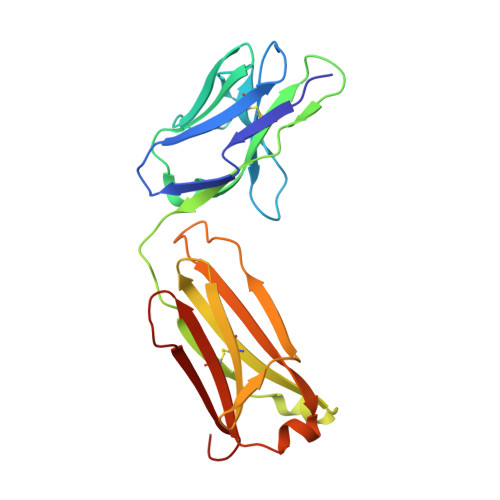
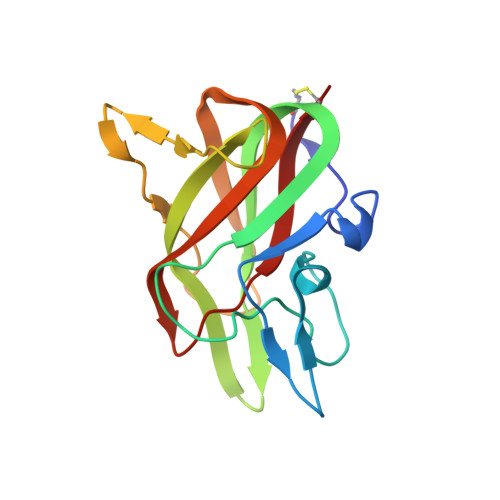
Structure of the Human Factor VIII C2 Domain in Complex with the 3E6 Inhibitory Antibody.
Wuerth, M.E., Cragerud, R.K., Clint Spiegel, P.(2015) Sci Rep 5: 17216-17216
- PubMed: 26598467
- DOI: https://doi.org/10.1038/srep17216
- Primary Citation of Related Structures:
4XZU - PubMed Abstract:
Blood coagulation factor VIII is a glycoprotein cofactor that is essential for the intrinsic pathway of the blood coagulation cascade. Inhibitory antibodies arise either spontaneously or in response to therapeutic infusion of functional factor VIII into hemophilia A patients, many of which are specific to the factor VIII C2 domain. The immune response is largely parsed into "classical" and "non-classical" inhibitory antibodies, which bind to opposing faces cooperatively. In this study, the 2.61 Å resolution structure of the C2 domain in complex with the antigen-binding fragment of the 3E6 classical inhibitory antibody is reported. The binding interface is largely conserved when aligned with the previously determined structure of the C2 domain in complex with two antibodies simultaneously. Further inspection of the B factors for the C2 domain in various X-ray crystal structures indicates that 3E6 antibody binding decreases the thermal motion behavior of surface loops in the C2 domain on the opposing face, thereby suggesting that cooperative antibody binding is a dynamic effect. Understanding the structural nature of the immune response to factor VIII following hemophilia A treatment will help lead to the development of better therapeutic reagents.
Organizational Affiliation:
Western Washington University, Department of Chemistry, 516 High Street, Bellingham, WA 98225-9150.