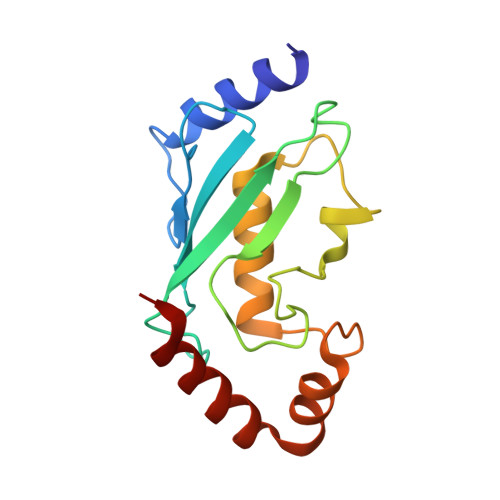
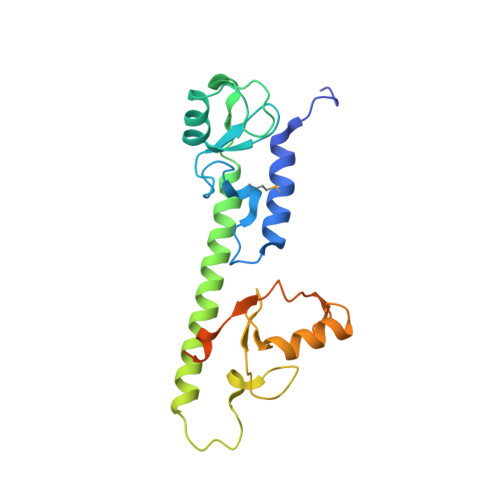
Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Quaile, A.T., Urbanus, M.L., Stogios, P.J., Nocek, B., Skarina, T., Ensminger, A.W., Savchenko, A.(2015) Structure 23: 1459-1469
- PubMed: 26146184
- DOI: https://doi.org/10.1016/j.str.2015.05.020
- Primary Citation of Related Structures:
4WZ0, 4WZ2, 4WZ3, 4XI1 - PubMed Abstract:
LubX is part of the large arsenal of effectors in Legionella pneumophila that are translocated into the host cytosol during infection. Despite such unique features as the presence of two U-box motifs and its targeting of another effector SidH, the molecular basis of LubX activity remains poorly understood. Here we show that the N terminus of LubX is able to activate an extended number of ubiquitin-conjugating (E2) enzymes including UBE2W, UBEL6, and all tested members of UBE2D and UBE2E families. Crystal structures of LubX alone and in complex with UBE2D2 revealed drastic molecular diversification between the two U-box domains, with only the N-terminal U-box retaining E2 recognition features typical for its eukaryotic counterparts. Extensive mutagenesis followed by functional screening in a yeast model system captured functionally important LubX residues including Arg121, critical for interactions with SidH. Combined, these data provide a new molecular insight into the function of this unique pathogenic factor.
Organizational Affiliation:
Department of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, ON M5S 3E5, Canada.