Structure of a TCR-Mimic Antibody with Target Predicts Pharmacogenetics.
Ataie, N., Xiang, J., Cheng, N., Brea, E.J., Lu, W., Scheinberg, D.A., Liu, C., Ng, H.L.(2016) J Mol Biol 428: 194-205
- PubMed: 26688548
- DOI: https://doi.org/10.1016/j.jmb.2015.12.002
- Primary Citation of Related Structures:
4WUU - PubMed Abstract:
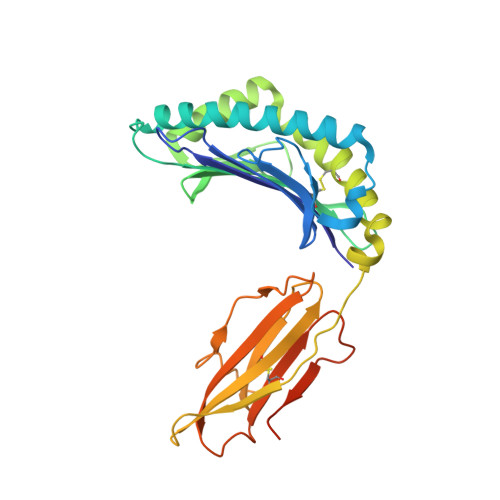
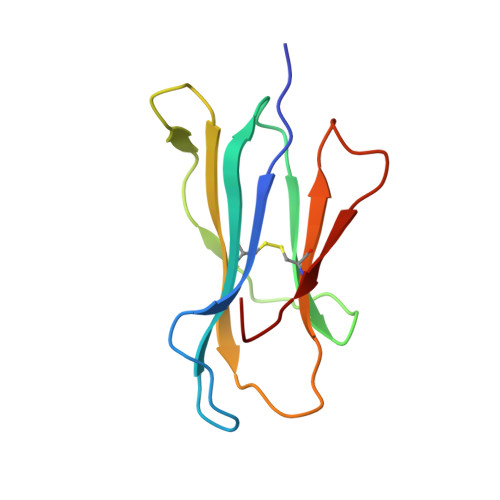
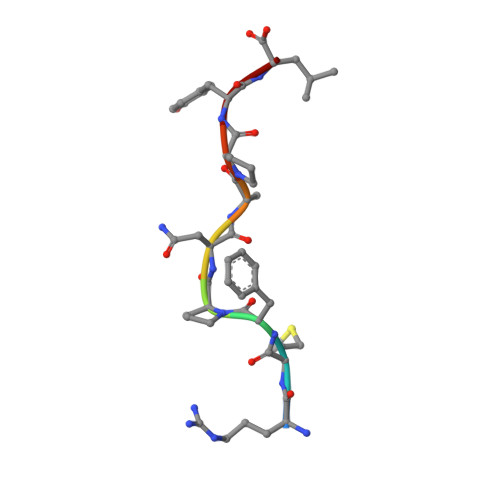
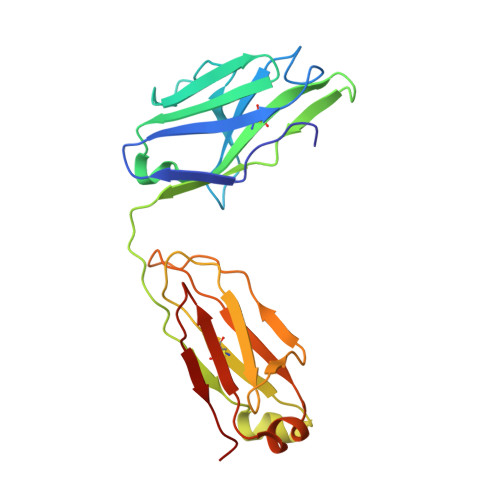
Antibody therapies currently target only extracellular antigens. A strategy to recognize intracellular antigens is to target peptides presented by immune HLA receptors. ESK1 is a human, T-cell receptor (TCR)-mimic antibody that binds with subnanomolar affinity to the RMF peptide from the intracellular Wilms tumor oncoprotein WT1 in complex with HLA-A*02:01. ESK1 is therapeutically effective in mouse models of WT1(+) human cancers. TCR-based therapies have been presumed to be restricted to one HLA subtype. The mechanism for the specificity and high affinity of ESK1 is unknown. We show in a crystal structure that ESK1 Fab binds to RMF/HLA-A*02:01 in a mode different from that of TCRs. From the structure, we predict and then experimentally confirm high-affinity binding with multiple other HLA-A*02 subtypes, broadening the potential patient pool for ESK1 therapy. Using the crystal structure, we also predict potential off-target binding that we experimentally confirm. Our results demonstrate how protein structure information can contribute to personalized immunotherapy.
Organizational Affiliation:
University of Hawaii at Manoa, Department of Chemistry, 2545 McCarthy Mall, Honolulu, HI 96822-2275, USA.