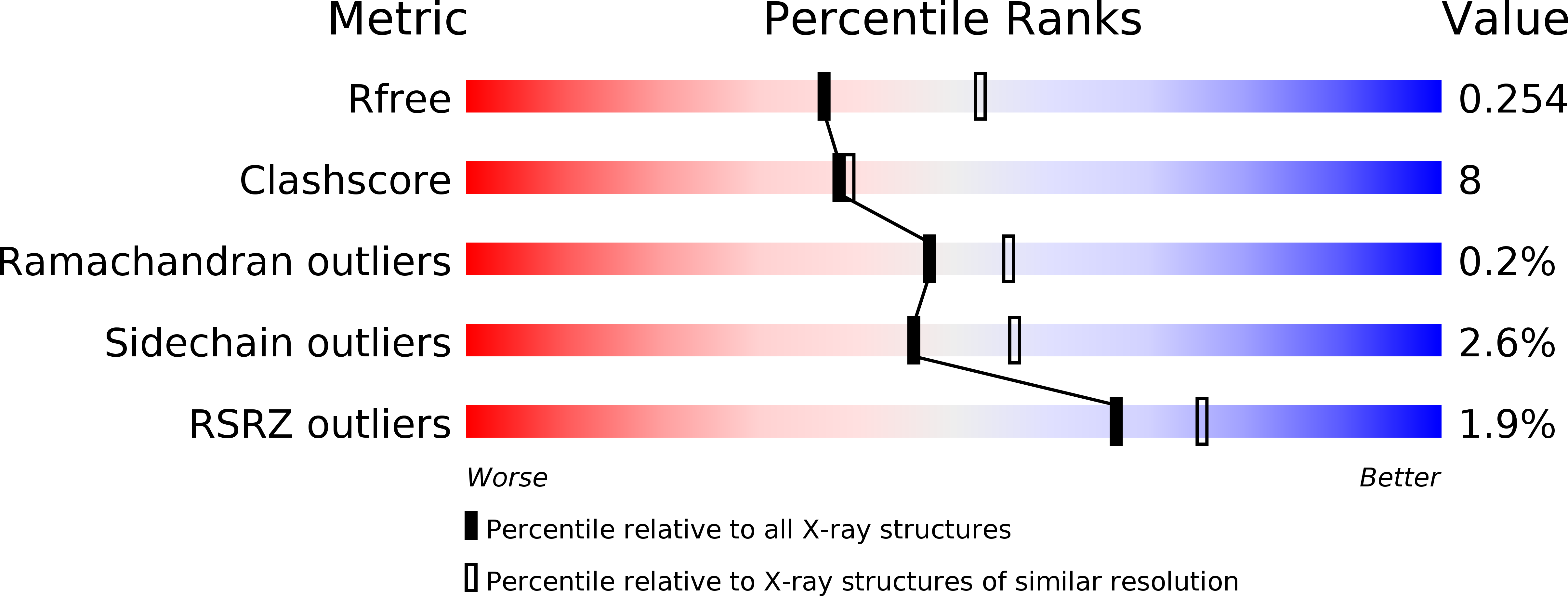
The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Zhang, W., Collinet, B., Perrochia, L., Durand, D., van Tilbeurgh, H.(2015) Nucleic Acids Res 43: 1804-1817
- PubMed: 25578970
- DOI: https://doi.org/10.1093/nar/gku1397
- Primary Citation of Related Structures:
4WQ4, 4WQ5, 4YDU - PubMed Abstract:
The essential and universal N(6)-threonylcarbamoyladenosine (t(6)A) modification at position 37 of ANN-decoding tRNAs plays a pivotal role in translational fidelity through enhancement of the cognate codon recognition and stabilization of the codon-anticodon interaction. In Escherichia coli, the YgjD (TsaD), YeaZ (TsaB), YjeE (TsaE) and YrdC (TsaC) proteins are necessary and sufficient for the in vitro biosynthesis of t(6)A, using tRNA, ATP, L-threonine and bicarbonate as substrates. YrdC synthesizes the short-lived L-threonylcarbamoyladenylate (TCA), and YgjD, YeaZ and YjeE cooperate to transfer the L-threonylcarbamoyl-moiety from TCA onto adenosine at position 37 of substrate tRNA. We determined the crystal structure of the heterodimer YgjD-YeaZ at 2.3 Å, revealing the presence of an unexpected molecule of ADP bound at an atypical site situated at the YgjD-YeaZ interface. We further showed that the ATPase activity of YjeE is strongly activated by the YgjD-YeaZ heterodimer. We established by binding experiments and SAXS data analysis that YgjD-YeaZ and YjeE form a compact ternary complex only in presence of ATP. The formation of the ternary YgjD-YeaZ-YjeE complex is required for the in vitro biosynthesis of t(6)A but not its ATPase activity.
Organizational Affiliation:
Institut de Biochimie et Biophysique Moléculaire et Cellulaire, UMR 8619, CNRS, Bâtiment 430, Université de Paris-Sud, 91405 Orsay Cedex, France.