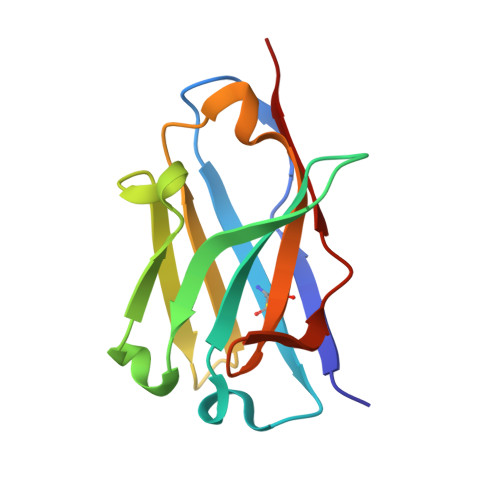
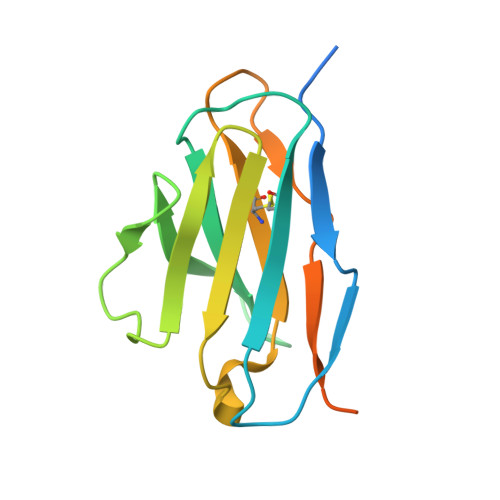
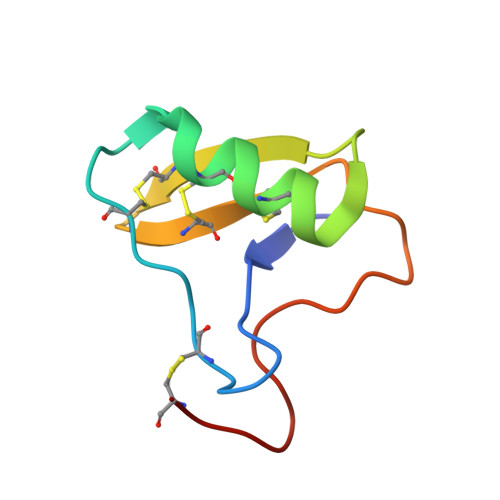
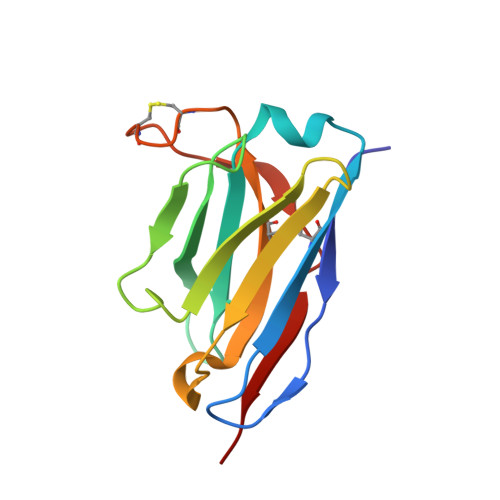
Optimal Neutralization of Centruroides Noxius Venom is Understood Through a Structural Complex between Two Antibody Fragments and the Cn2 Toxin.
Riano-Umbarila, L., Ledezma-Candanoza, L.M., Serrano-Posada, H., Fernandez-Taboada, G., Olamendi-Portugal, T., Rojas-Trejo, S., Gomez-Ramirez, I.V., Rudino-Pinera, E., Possani, L.D., Becerril, B.(2016) J Biol Chem 291: 1619
- PubMed: 26589800
- DOI: https://doi.org/10.1074/jbc.M115.685297
- Primary Citation of Related Structures:
4V1D - PubMed Abstract:
The current trend of using recombinant antibody fragments in research to develop novel antidotes against scorpion stings has achieved excellent results. The polyclonal character of commercial antivenoms, obtained through the immunization of animals and which contain several neutralizing antibodies that recognize different epitopes on the toxins, guarantees the neutralization of the venoms. To avoid the use of animals, we aimed to develop an equivalent recombinant antivenom composed of a few neutralizing single chain antibody fragments (scFvs) that bind to two different epitopes on the scorpion toxins. In this study, we obtained scFv RU1 derived from scFv C1. RU1 showed a good capacity to neutralize the Cn2 toxin and whole venom of the scorpion Centruroides noxius. Previously, we had produced scFv LR, obtained from a different parental fragment (scFv 3F). LR also showed a similar neutralizing capacity. The simultaneous administration of both scFvs resulted in improved protection, which was translated as a rapid recovery of previously poisoned animals. The crystallographic structure of the ternary complex scFv LR-Cn2-scFv RU1 allowed us to identify the areas of interaction of both scFvs with the toxin, which correspond to non-overlapping sites. The epitope recognized by scFv RU1 seems to be related to a greater efficiency in the neutralization of the whole venom. In addition, the structural analysis of the complex helped us to explain the cross-reactivity of these scFvs and how they neutralize the venom.
Organizational Affiliation:
From the Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Apartado Postal 510-3, Cuernavaca, Morelos 62250, México and.