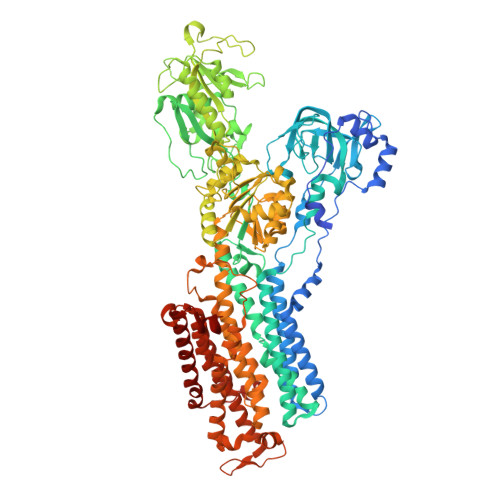
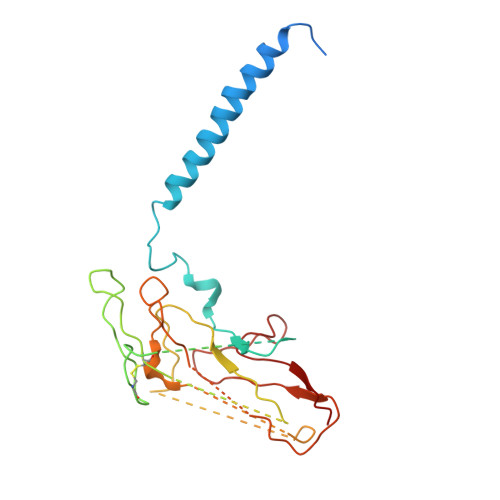
Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
Abe, K., Tani, K., Fujiyoshi, Y.(2014) J Biol Chem 289: 30590
- PubMed: 25231997
- DOI: https://doi.org/10.1074/jbc.M114.584623
- Primary Citation of Related Structures:
4UX1, 4UX2 - PubMed Abstract:
Gastric H(+),K(+)-ATPase, an ATP-driven proton pump responsible for gastric acidification, is a molecular target for anti-ulcer drugs. Here we show its cryo-electron microscopy (EM) structure in an E2P analog state, bound to magnesium fluoride (MgF), and its K(+)-competitive antagonist SCH28080, determined at 7 Å resolution by electron crystallography of two-dimensional crystals. Systematic comparison with other E2P-related cryo-EM structures revealed that the molecular conformation in the (SCH)E2·MgF state is remarkably distinguishable. Although the azimuthal position of the A domain of the (SCH)E2·MgF state is similar to that in the E2·AlF (aluminum fluoride) state, in which the transmembrane luminal gate is closed, the arrangement of transmembrane helices in the (SCH)E2·MgF state shows a luminal-open conformation imposed on by bound SCH28080 at its luminal cavity, based on observations of the structure in the SCH28080-bound E2·BeF (beryllium fluoride) state. The molecular conformation of the (SCH)E2·MgF state thus represents a mixed overall structure in which its cytoplasmic and luminal half appear to be independently modulated by a phosphate analog and an antagonist bound to the respective parts of the enzyme. Comparison of the molecular conformations revealed that the linker region connecting the A domain and the transmembrane helix 2 (A-M2 linker) mediates the regulation of luminal gating. The mechanistic rationale underlying luminal gating observed in H(+),K(+)-ATPase is consistent with that observed in sarcoplasmic reticulum Ca(2+)-ATPase and other P-type ATPases and is most likely conserved for the P-type ATPase family in general.
Organizational Affiliation:
Cellular and Structural Physiology Institute and Nagoya University, Nagoya 464-8601, Japan; Graduate School of Pharmaceutical Science, Nagoya University, Nagoya 464-8601, Japan. Electronic address: kabe@cespi.nagoya-u.ac.jp.