The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with pyruvic acid
Tan, K., Li, H., Jedrzejczak, R., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Amino acid/amide ABC transporter substrate-binding protein, HAAT family | 395 | Trichormus variabilis ATCC 29413 | Mutation(s): 0 Gene Names: Ava_0465 | 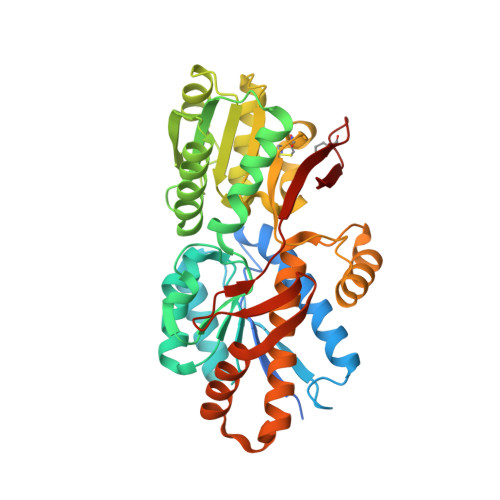 | |
UniProt | |||||
Find proteins for Q3MFZ5 (Trichormus variabilis (strain ATCC 29413 / PCC 7937)) Explore Q3MFZ5 Go to UniProtKB: Q3MFZ5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q3MFZ5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PYR Query on PYR | C [auth A], F [auth B] | PYRUVIC ACID C3 H4 O3 LCTONWCANYUPML-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | E [auth A] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| MG Query on MG | D [auth A], G [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 98.525 | α = 90 |
| b = 100.588 | β = 90 |
| c = 150.602 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| SHELXD | phasing |
| MLPHARE | phasing |
| DM | model building |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| DM | phasing |