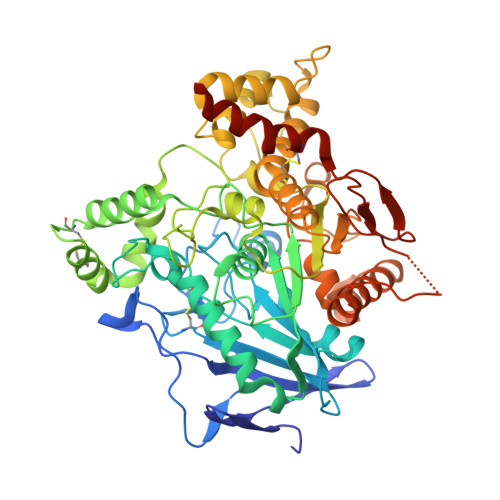
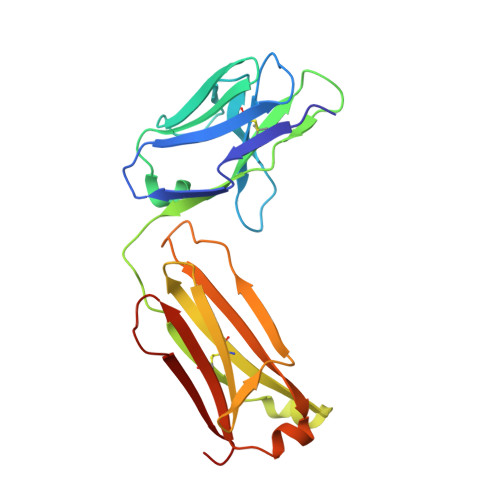
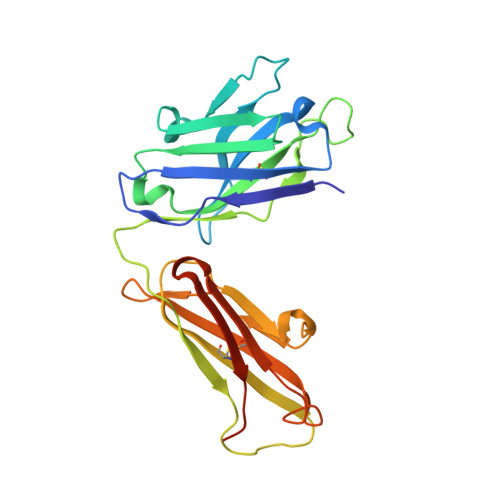
Crystal Structure of Snake Venom Acetylcholinesterase in Complex with Inhibitory Antibody Fragment Fab410 Bound at the Peripheral Site: EVIDENCE FOR OPEN AND CLOSED STATES OF A BACK DOOR CHANNEL.
Bourne, Y., Renault, L., Marchot, P.(2015) J Biol Chem 290: 1522-1535
- PubMed: 25411244
- DOI: https://doi.org/10.1074/jbc.M114.603902
- Primary Citation of Related Structures:
4QWW - PubMed Abstract:
The acetylcholinesterase found in the venom of Bungarus fasciatus (BfAChE) is produced as a soluble, non-amphiphilic monomer with a canonical catalytic domain but a distinct C terminus compared with the other vertebrate enzymes. Moreover, the peripheral anionic site of BfAChE, a surface site located at the active site gorge entrance, bears two substitutions altering sensitivity to cationic inhibitors. Antibody Elec410, generated against Electrophorus electricus acetylcholinesterase (EeAChE), inhibits EeAChE and BfAChE by binding to their peripheral sites. However, both complexes retain significant residual catalytic activity, suggesting incomplete gorge occlusion by bound antibody and/or high frequency back door opening. To explore a novel acetylcholinesterase species, ascertain the molecular bases of inhibition by Elec410, and document the determinants and mechanisms for back door opening, we solved a 2.7-Å resolution crystal structure of natural BfAChE in complex with antibody fragment Fab410. Crystalline BfAChE forms the canonical dimer found in all acetylcholinesterase structures. Equally represented open and closed states of a back door channel, associated with alternate positions of a tyrosine phenol ring at the active site base, coexist in each subunit. At the BfAChE molecular surface, Fab410 is seated on the long Ω-loop between two N-glycan chains and partially occludes the gorge entrance, a position that fully reflects the available mutagenesis and biochemical data. Experimentally based flexible molecular docking supports a similar Fab410 binding mode onto the EeAChE antigen. These data document the molecular and dynamic peculiarities of BfAChE with high frequency back door opening, and the mode of action of Elec410 as one of the largest peptidic inhibitors targeting the acetylcholinesterase peripheral site.
Organizational Affiliation:
From Aix-Marseille Université, Architecture et Fonction des Macromolécules Biologiques, campus Luminy, 13228 Marseille cedex 09, France, CNRS, Architecture et Fonction des Macromolécules Biologiques, campus Luminy, 13228 Marseille cedex 09, France, and.