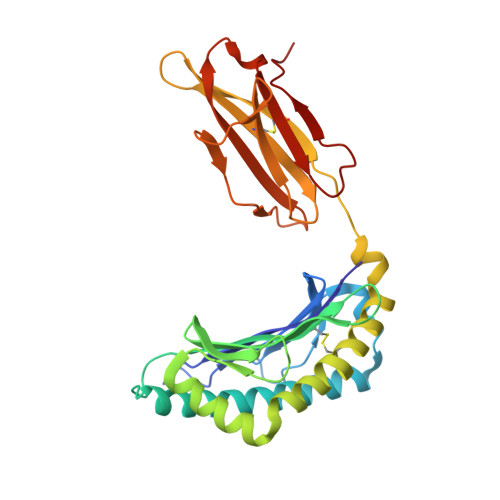
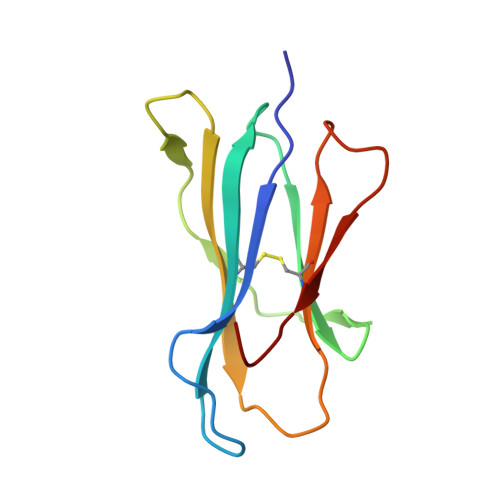
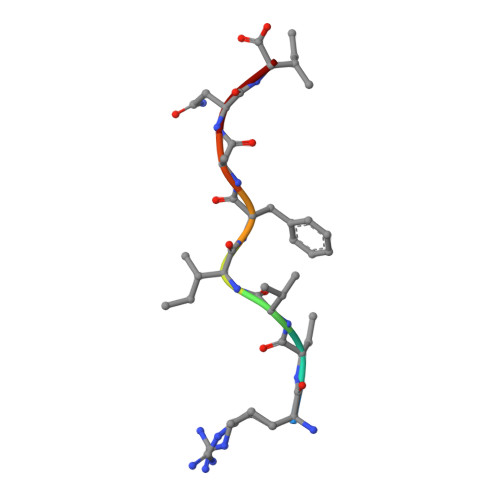
Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes.
Trujillo, J.A., Gras, S., Twist, K.A., Croft, N.P., Channappanavar, R., Rossjohn, J., Purcell, A.W., Perlman, S.(2014) J Immunol 192: 5245-5256
- PubMed: 24795457
- DOI: https://doi.org/10.4049/jimmunol.1400111
- Primary Citation of Related Structures:
4PV8, 4PV9 - PubMed Abstract:
Peptides that bind poorly to MHC class I molecules often elicit low-functional avidity T cell responses. Peptide modification by altering the anchor residue facilitates increased binding affinity and may elicit T cells with increased functional avidity toward the native epitope ("heteroclitic"). This augmented MHC binding is likely to increase the half-life and surface density of the heteroclitic complex, but precisely how this enhanced T cell response occurs in vivo is not known. Furthermore, the ideal heteroclitic epitope will elicit T cell responses that completely cross-react with the native epitope, maximizing protection and minimizing undesirable off-target effects. Such epitopes have been difficult to identify. In this study, using mice infected with a murine coronavirus that encodes epitopes that elicit high (S510, CSLWNGPHL)- and low (S598, RCQIFANI)-functional avidity responses, we show that increased expression of peptide S598 but not S510 generated T cells with enhanced functional avidity. Thus, immune responses can be augmented toward T cell epitopes with low functional avidity by increasing Ag density. We also identified a heteroclitic epitope (RCVIFANI) that elicited a T cell response with nearly complete cross-reactivity with native epitope and demonstrated increased MHC/peptide abundance compared with native S598. Structural and thermal melt analyses indicated that the Q600V substitution enhanced stability of the peptide/MHC complex without greatly altering the antigenic surface, resulting in highly cross-reactive T cell responses. Our data highlight that increased peptide/MHC complex display contributes to heteroclitic epitope efficacy and describe parameters for maximizing immune responses that cross-react with the native epitope.
Organizational Affiliation:
Interdisciplinary Program in Immunology, University of Iowa, Iowa City, IA 52242;