The cellular redox environment alters antigen presentation.
Trujillo, J.A., Croft, N.P., Dudek, N.L., Channappanavar, R., Theodossis, A., Webb, A.I., Dunstone, M.A., Illing, P.T., Butler, N.S., Fett, C., Tscharke, D.C., Rossjohn, J., Perlman, S., Purcell, A.W.(2014) J Biol Chem 289: 27979-27991
- PubMed: 25135637
- DOI: https://doi.org/10.1074/jbc.M114.573402
- Primary Citation of Related Structures:
4PG2 - PubMed Abstract:
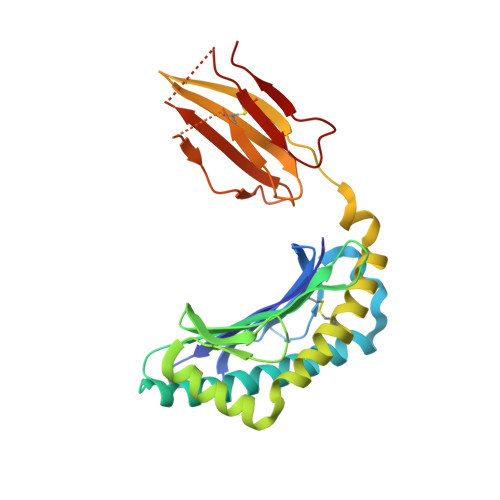
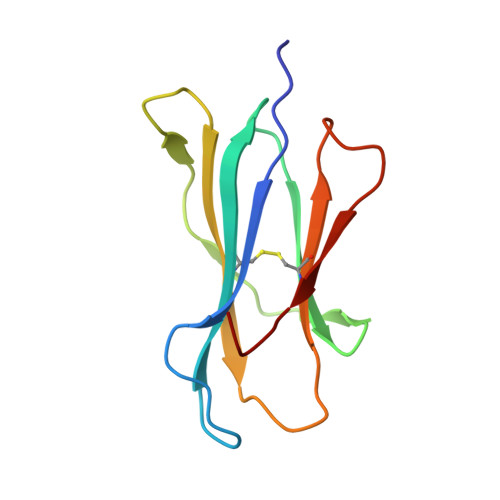
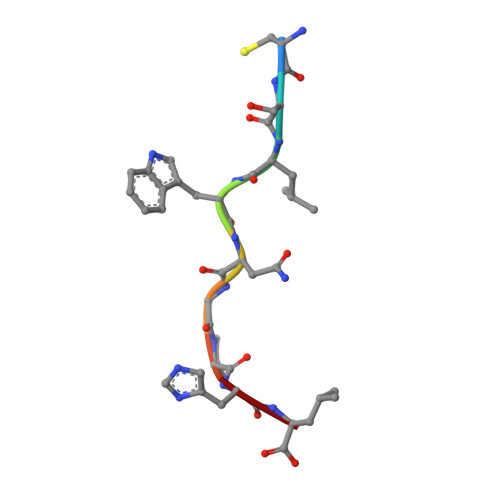
Cysteine-containing peptides represent an important class of T cell epitopes, yet their prevalence remains underestimated. We have established and interrogated a database of around 70,000 naturally processed MHC-bound peptides and demonstrate that cysteine-containing peptides are presented on the surface of cells in an MHC allomorph-dependent manner and comprise on average 5-10% of the immunopeptidome. A significant proportion of these peptides are oxidatively modified, most commonly through covalent linkage with the antioxidant glutathione. Unlike some of the previously reported cysteine-based modifications, this represents a true physiological alteration of cysteine residues. Furthermore, our results suggest that alterations in the cellular redox state induced by viral infection are communicated to the immune system through the presentation of S-glutathionylated viral peptides, resulting in altered T cell recognition. Our data provide a structural basis for how the glutathione modification alters recognition by virus-specific T cells. Collectively, these results suggest that oxidative stress represents a mechanism for modulating the virus-specific T cell response.
Organizational Affiliation:
From the Department of Microbiology and the Interdisciplinary Program in Immunology, University of Iowa, Iowa City, Iowa 52242.