The intramolecular disulfide-stapled structure of laterosporulin, a class IId bacteriocin, conceals a human defensin-like structural module.
Singh, P.K., Solanki, V., Sharma, S., Thakur, K.G., Krishnan, B., Korpole, S.(2015) FEBS J 282: 203-214
- PubMed: 25345978
- DOI: https://doi.org/10.1111/febs.13129
- Primary Citation of Related Structures:
4OZK - PubMed Abstract:
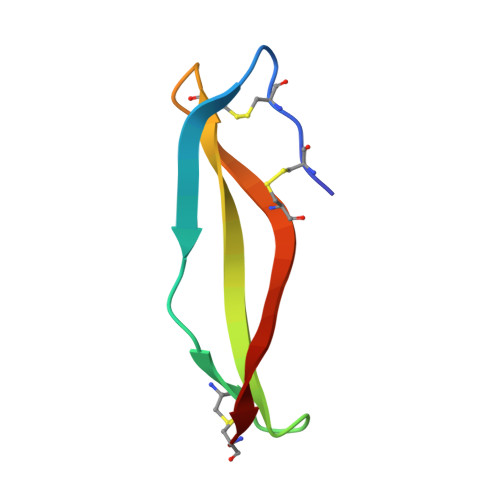
The growing emergence of antibiotic-resistant bacteria has led to the exploration of naturally occurring defense peptides as antimicrobials. In this study, we found that laterosporulin (LS), a class IId bacteriocin, effectively kills active and nonmultiplying cells of both Gram-positive and Gram-negative bacteria. Fluorescence and electron microscopy suggest that growth inhibition occurs because of increased membrane permeability. The crystal structure of LS at 2.0 Å resolution reveals an all-β conformation of this peptide, with four β-strands forming a twisted β-sheet. All six intrinsic cysteines are intramolecularly disulfide-bonded, with two disulfides constraining the N terminus of the peptide and the third disulfide crosslinking the extreme C terminus, resulting in the formation of a closed structure. The significance of disulfides in maintaining the in-solution peptide structure was confirmed by CD and fluorescence analyses. Despite a low overall sequence similarity, LS has disulfide connectivity [C(I)-C(V), C(II)-C(IV), and C(III)-C(VI)] like that of β-defensins and a striking architectural similarity with α-defensins. Therefore LS presents a missing link between bacteriocins and mammalian defensins, and is also a potential antimicrobial lead, in particular against nonmultiplying bacteria.
Organizational Affiliation:
Microbial Type Culture Collection & Gene Bank, CSIR-Institute of Microbial Technology, Chandigarh, India.