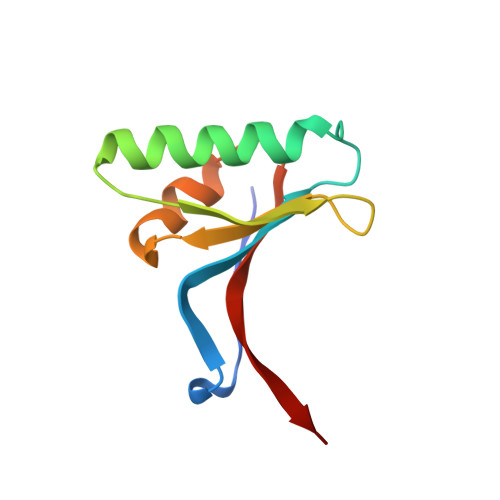
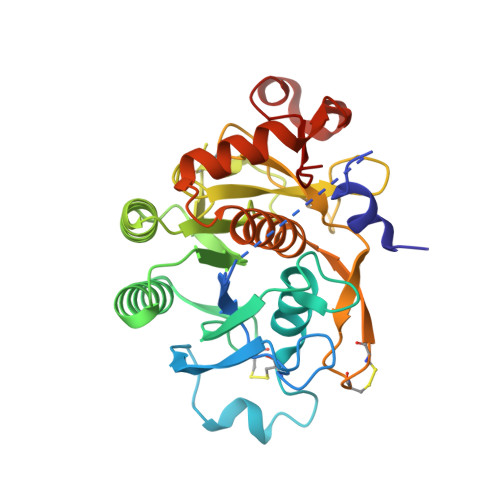
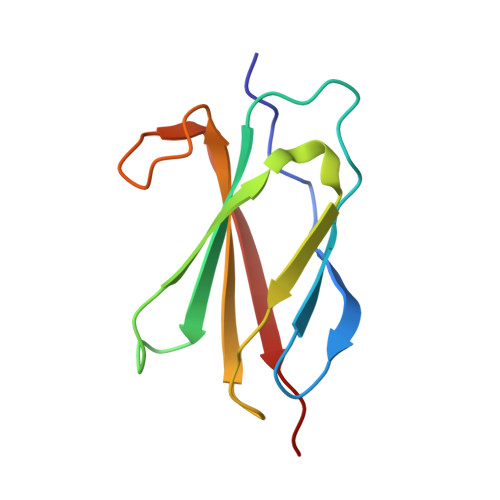
Pharmacologic Profile of the Adnectin BMS-962476, a Small Protein Biologic Alternative to PCSK9 Antibodies for Low-Density Lipoprotein Lowering.
Mitchell, T., Chao, G., Sitkoff, D., Lo, F., Monshizadegan, H., Meyers, D., Low, S., Russo, K., DiBella, R., Denhez, F., Gao, M., Myers, J., Duke, G., Witmer, M., Miao, B., Ho, S.P., Khan, J., Parker, R.A.(2014) J Pharmacol Exp Ther 350: 412-424
- PubMed: 24917546
- DOI: https://doi.org/10.1124/jpet.114.214221
- Primary Citation of Related Structures:
4OV6 - PubMed Abstract:
Proprotein convertase subtilisin kexin-9 (PCSK9) is an important pharmacological target for decreasing low-density lipoprotein (LDL) in cardiovascular disease, although seemingly inaccessible to small molecule approaches. Compared with therapeutic IgG antibodies currently in development, targeting circulating PCSK9 with smaller molecular scaffolds could offer different profiles and reduced dose burdens. This inspired genesis of PCSK9-binding Adnectins, a protein family derived from human fibronectin-10th-type III-domain and engineered for high-affinity target binding. BMS-962476, an ∼11-kDa polypeptide conjugated to polyethylene glycol to enhance pharmacokinetics, binds with subnanomolar affinity to human. The X-ray cocrystal structure of PCSK9 with a progenitor Adnectin shows ∼910 Å(2) of PCSK9 surface covered next to the LDL receptor binding site, largely by residues of a single loop of the Adnectin. In hypercholesterolemic, overexpressing human PCSK9 transgenic mice, BMS-962476 rapidly lowered cholesterol and free PCSK9 levels. In genomic transgenic mice, BMS-962476 potently reduced free human PCSK9 (ED50 ∼0.01 mg/kg) followed by ∼2-fold increases in total PCSK9 before return to baseline. Treatment of cynomolgus monkeys with BMS-962476 rapidly suppressed free PCSK9 >99% and LDL-cholesterol ∼55% with subsequent 6-fold increase in total PCSK9, suggesting reduced clearance of circulating complex. Liver sterol response genes were consequently downregulated, following which LDL and total PCSK9 returned to baseline. These studies highlight the rapid dynamics of PCSK9 control over LDL and liver cholesterol metabolism and characterize BMS-962476 as a potent and efficacious PCSK9 inhibitor.
Organizational Affiliation:
Molecular Discovery Technologies (T.M., G.C., D.S., S.L., K.R., R.D., F.D., M.G., J.M., G.D., M.W., J.K.), Applied Genomics (S.P.H.), and Cardiovascular Discovery Biology (F.L., H.M., D.M., B.M., R.A.P.), Bristol-Myers Squibb Research and Development, Princeton, New Jersey rex_a_parker@msn.com tracy.mitchell@bms.com.