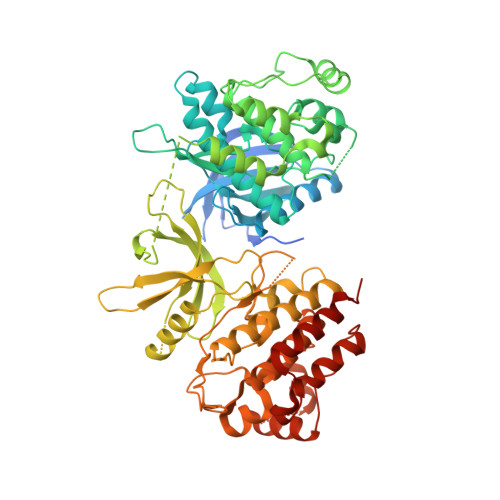
Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition.
Lupardus, P.J., Ultsch, M., Wallweber, H., Bir Kohli, P., Johnson, A.R., Eigenbrot, C.(2014) Proc Natl Acad Sci U S A 111: 8025-8030
- PubMed: 24843152
- DOI: https://doi.org/10.1073/pnas.1401180111
- Primary Citation of Related Structures:
4OLI - PubMed Abstract:
Janus kinases (JAKs) are receptor-associated multidomain tyrosine kinases that act downstream of many cytokines and interferons. JAK kinase activity is regulated by the adjacent pseudokinase domain via an unknown mechanism. Here, we report the 2.8-Å structure of the two-domain pseudokinase-kinase module from the JAK family member TYK2 in its autoinhibited form. We find that the pseudokinase and kinase interact near the kinase active site and that most reported mutations in cancer-associated JAK alleles cluster in or near this interface. Mutation of residues near the TYK2 interface that are analogous to those in cancer-associated JAK alleles, including the V617F and "exon 12" JAK2 mutations, results in increased kinase activity in vitro. These data indicate that JAK pseudokinases are autoinhibitory domains that hold the kinase domain inactive until receptor dimerization stimulates transition to an active state.
Organizational Affiliation:
Departments of Structural Biology and lupardus.patrick@gene.com eigenbrot.c@gene.com.